4K39
 
 | | Native anSMEcpe with bound AdoMet and Cp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, Cp18Cys peptide, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K38
 
 | | Native anSMEcpe with bound AdoMet and Kp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K37
 
 | | Native anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K36
 
 | | His6 tagged anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYF
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYE
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | BROMIDE ION, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYD
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, BROMIDE ION, CHAPSO, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JY9
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JY8
 
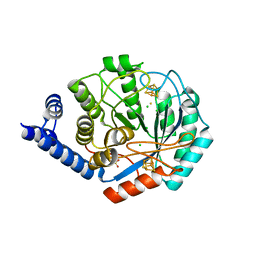 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHLORIDE ION, FEFE-HYDROGENASE MATURASE, HYDROSULFURIC ACID, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JXC
 
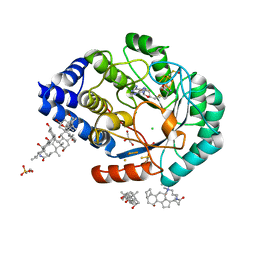 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JC0
 
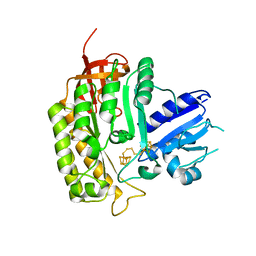 | | Crystal structure of Thermotoga maritima holo RimO in complex with pentasulfide, Northeast Structural Genomics Consortium Target VR77 | | Descriptor: | IRON/SULFUR PENTA-SULFIDE CONNECTED CLUSTERS, Ribosomal protein S12 methylthiotransferase RimO | | Authors: | Forouhar, F, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases.
Nat.Chem.Biol., 9, 2013
|
|
3T7V
 
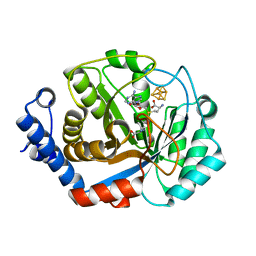 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3RFA
 
 | | X-ray structure of RlmN from Escherichia coli in complex with S-adenosylmethionine | | Descriptor: | IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N, S-ADENOSYLMETHIONINE | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3RF9
 
 | | X-ray structure of RlmN from Escherichia coli | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3IIZ
 
 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with S-adenosyl-L-methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3IIX
 
 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with methionine and 5'deoxyadenosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBONATE ION, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3CIX
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima in complex with thiocyanate | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
3CB8
 
 | |
3CAN
 
 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
3C8F
 
 | |
2Z2U
 
 | | Crystal structure of archaeal TYW1 | | Descriptor: | UPF0026 protein MJ0257 | | Authors: | Suzuki, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2007-05-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA
J.Mol.Biol., 372, 2007
|
|
2YX0
 
 | | Crystal structure of P. horikoshii TYW1 | | Descriptor: | radical sam enzyme | | Authors: | Goto-Ito, S, Ishii, R, Ito, T, Shibata, R, Fusatomi, E, Sekine, S, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QGQ
 
 | | Crystal structure of TM_1862 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR77 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Protein TM_1862 | | Authors: | Forouhar, F, Neely, H, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Post-translational Modification of Ribosomal Proteins: STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF RimO FROM THERMOTOGA MARITIMA, A RADICAL S-ADENOSYLMETHIONINE METHYLTHIOTRANSFERASE.
J.Biol.Chem., 285, 2010
|
|
2FB3
 
 | | Structure of MoaA in complex with 5'-GTP | | Descriptor: | 5'-DEOXYADENOSINE, GUANOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
