4WPO
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the pre-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-20 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQU
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G trapped by the antibiotic dityromycin | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4D61
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
5AFI
 
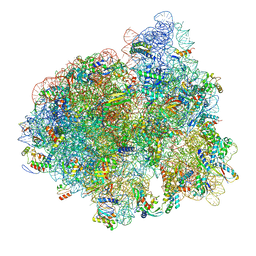 | | 2.9A Structure of E. coli ribosome-EF-TU complex by cs-corrected cryo-EM | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Konevega, A.L, Bock, L.V, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the E. coli ribosome-EF-Tu complex at <3 angstrom resolution by Cs-corrected cryo-EM.
Nature, 520, 2015
|
|
4PC1
 
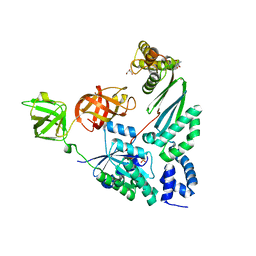 | | Elongation Factor Tu:Ts complex with a bound phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC7
 
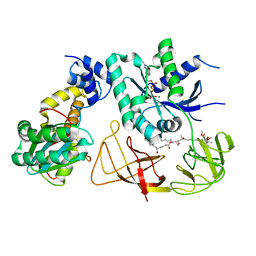 | | Elongation factor Tu:Ts complex in a near GTP conformation. | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6003 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC3
 
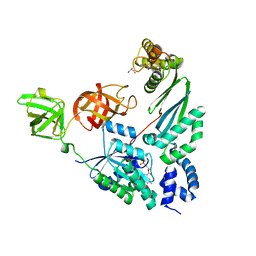 | | Elongation factor Tu:Ts complex with partially bound GDP | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8313 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC2
 
 | | Elongation factor Tu:Ts complex with a bound GDP | | Descriptor: | CHLORIDE ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC6
 
 | | Elongation factor Tu:Ts complex with bound GDPNP | | Descriptor: | Elongation factor Ts, Elongation factor Tu, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
3J9Z
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
3JA1
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
4ZCM
 
 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with ppGpp | | Descriptor: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCL
 
 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with GDP | | Descriptor: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCI
 
 | | Crystal Structure of Escherichia coli GTPase BipA/TypA | | Descriptor: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.627 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
3JAP
 
 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
5A9Y
 
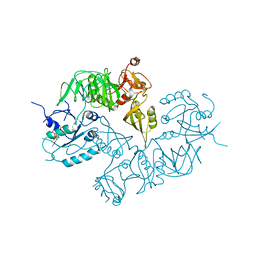 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
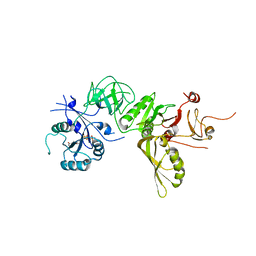 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
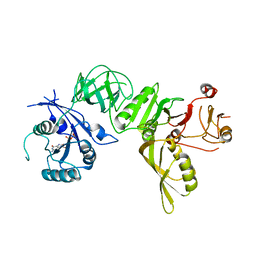 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
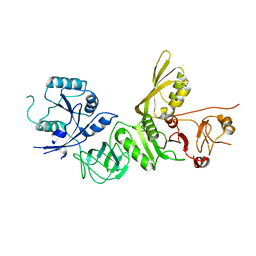 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R71
 
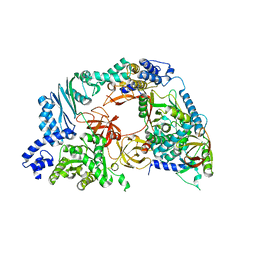 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
4ZU9
 
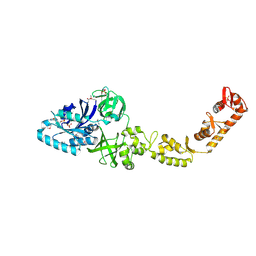 | | Crystal structure of bacterial selenocysteine-specific elongation factor EF-Sec | | Descriptor: | CYSTEINE, Elongation factor SelB, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S. | | Deposit date: | 2015-05-15 | | Release date: | 2015-09-23 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB
Nucleic Acids Res., 43, 2015
|
|
3JB9
 
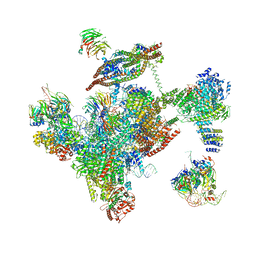 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
5AA0
 
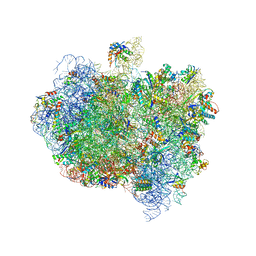 | | Complex of Thermous thermophilus ribosome (A-and P-site tRNA) bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9Z
 
 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
