5A9V
 
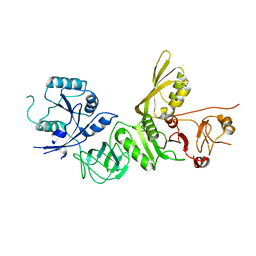 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3JB9
 
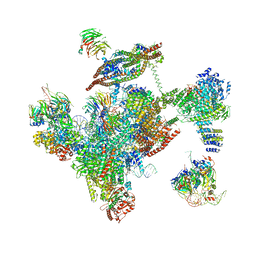 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCJ
 
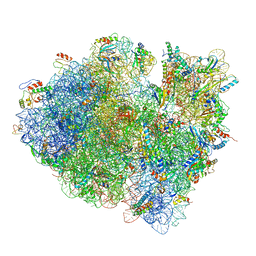 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3J4J
 
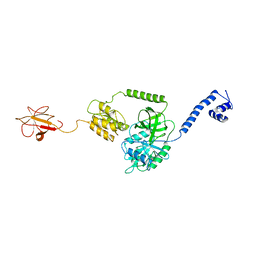 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JAP
 
 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3JCN
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association: Initiation Complex I | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3JA1
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
4PC3
 
 | | Elongation factor Tu:Ts complex with partially bound GDP | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8313 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC7
 
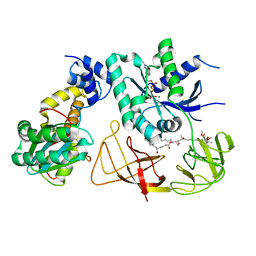 | | Elongation factor Tu:Ts complex in a near GTP conformation. | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6003 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4P3Y
 
 | |
4PC1
 
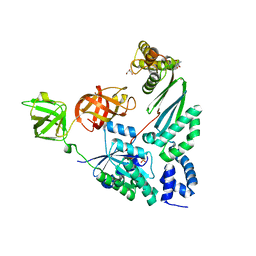 | | Elongation Factor Tu:Ts complex with a bound phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC6
 
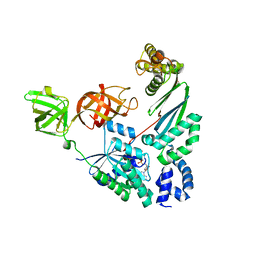 | | Elongation factor Tu:Ts complex with bound GDPNP | | Descriptor: | Elongation factor Ts, Elongation factor Tu, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
4PC2
 
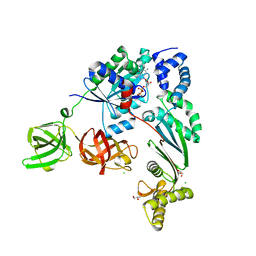 | | Elongation factor Tu:Ts complex with a bound GDP | | Descriptor: | CHLORIDE ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
3AGP
 
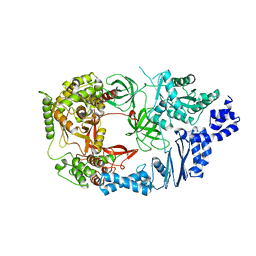 | | Structure of viral polymerase form I | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AGQ
 
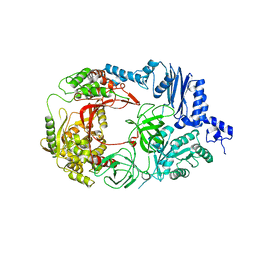 | | Structure of viral polymerase form II | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, LINKER, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AVT
 
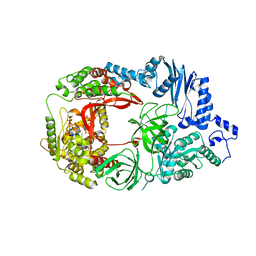 | | Structure of viral RNA polymerase complex 1 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVU
 
 | | Structure of viral RNA polymerase complex 2 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVW
 
 | | Structure of viral RNA polymerase complex 4 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVV
 
 | | Structure of viral RNA polymerase complex 3 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.119 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVX
 
 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVY
 
 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3B82
 
 | | Structure of the eEF2-ExoA(E546H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B8H
 
 | | Structure of the eEF2-ExoA(E546A)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-11-01 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B78
 
 | | Structure of the eEF2-ExoA(R551H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-30 | | Release date: | 2008-06-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3CB4
 
 | | The Crystal Structure of LepA | | Descriptor: | GTP-binding protein lepA | | Authors: | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
