4ID9
 
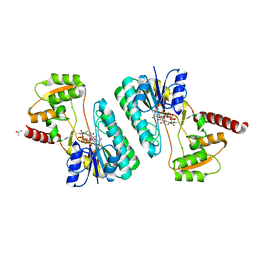 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1 | | 分子名称: | ALANINE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-12-12 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1
To be Published
|
|
3EHE
 
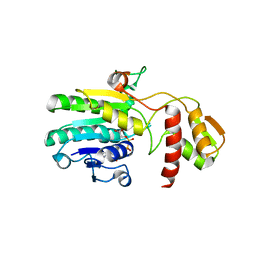 | |
3GPI
 
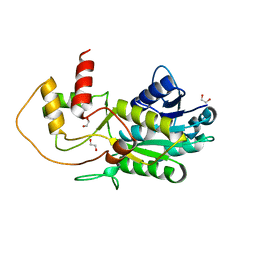 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | 分子名称: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | 著者 | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-23 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
4LIS
 
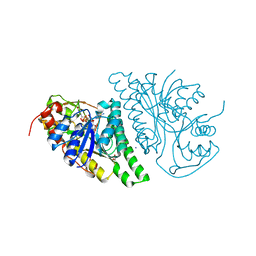 | | Crystal Structure of UDP-galactose-4-epimerase from Aspergillus nidulans | | 分子名称: | GLYCEROL, IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Dalrymple, S.A, Ko, J, Sheoran, I, Kaminskyj, S.G.W, Sanders, D.A.R. | | 登録日 | 2013-07-03 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Elucidation of Substrate Specificity in Aspergillus nidulans UDP-Galactose-4-Epimerase.
Plos One, 8, 2013
|
|
4LW8
 
 | |
3HFS
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | 分子名称: | Anthocyanidin reductase, CHLORIDE ION | | 著者 | Gargouri, M, Mauge, C, Langlois d'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | 登録日 | 2009-05-12 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ICP
 
 | |
5EMH
 
 | |
3IUS
 
 | | The structure of a functionally unknown conserved protein from Silicibacter pomeroyi DSS | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, uncharacterized conserved protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-31 | | 公開日 | 2009-10-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | The structure of a functionally unknown conserved protein from Silicibacter pomeroyi DSS
To be Published
|
|
4WOK
 
 | |
1A9Y
 
 | |
1A9Z
 
 | |
5GMO
 
 | | X-ray structure of carbonyl reductase SsCR | | 分子名称: | Protein induced by osmotic stress | | 著者 | Shang, Y.P, Yu, H.L, Xu, J.H. | | 登録日 | 2016-07-14 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Efficient Synthesis of (R)-2-Chloro-1-(2,4-dichlorophenyl)ethanol with a Ketoreductase from Scheffersomyces stipitis CBS 6045
Adv.Synth.Catal., 359, 2017
|
|
3KO8
 
 | | Crystal Structure of UDP-galactose 4-epimerase | | 分子名称: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Sakuraba, H, Kawai, T, Yoneda, K, Ohshima, T. | | 登録日 | 2009-11-13 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of UDP-galactose 4-epimerase from the hyperthermophilic archaeon Pyrobaculum calidifontis
Arch.Biochem.Biophys., 512, 2011
|
|
1BXK
 
 | | DTDP-GLUCOSE 4,6-DEHYDRATASE FROM E. COLI | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) | | 著者 | Thoden, J.B, Hegeman, A.D, Frey, P.A, Holden, H.M. | | 登録日 | 1998-10-05 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Structure of Dtdp-Glucose 4,6-Dehydratase from E. Coli
Protein Sci.
|
|
1BSV
 
 | |
1BWS
 
 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | 著者 | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | 登録日 | 1998-09-25 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
5GY7
 
 | | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis. | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NITRATE ION, ... | | 著者 | Singh, N, Tiwari, P, Phulera, S, Dixit, A, Choudhury, D. | | 登録日 | 2016-09-21 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis.
To Be Published
|
|
4YRB
 
 | | mouse TDH mutant R180K with NAD+ bound | | 分子名称: | L-threonine 3-dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | He, C, Li, F. | | 登録日 | 2015-03-14 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
4YRD
 
 | | Crystal structure of CapF with inhibitor 3-isopropenyl-tropolone | | 分子名称: | 2-hydroxy-3-(prop-1-en-2-yl)cyclohepta-2,4,6-trien-1-one, Capsular polysaccharide synthesis enzyme Cap5F, ZINC ION | | 著者 | Nakano, K, Chigira, T, Miyafusa, T, Nagatoishi, S, Caaveiro, J.M.M, Tsumoto, K. | | 登録日 | 2015-03-15 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Discovery and characterization of natural tropolones as inhibitors of the antibacterial target CapF from Staphylococcus aureus.
Sci Rep, 5, 2015
|
|
4YRA
 
 | | mouse TDH in the apo form | | 分子名称: | L-threonine 3-dehydrogenase, mitochondrial | | 著者 | He, C, Li, F. | | 登録日 | 2015-03-14 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
3LU1
 
 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | 分子名称: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | 著者 | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | 登録日 | 2010-02-16 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
3M2P
 
 | |
4QTZ
 
 | |
4R1T
 
 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | 分子名称: | cinnamoyl CoA reductase, molecular iodine | | 著者 | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
