5WWV
 
 | | Crystal structure of porcine kidney D-amino acid oxidase mutant (I230A/R283G) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-(4-chlorophenyl)-phenyl-methanamine, D-amino-acid oxidase, ... | | Authors: | Motojima, F, Yasukawa, K, Ohno, A, Asano, Y. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tailoring D-amino acid oxidase from the pid kidney to R-stereoselective amine oxidase and its use in the deracemization of 4-chlorobenzhydrylamine
To Be Published
|
|
5WX2
 
 | | Crystal structure of porcine kidney D-amino acid oxidase mutant (I230A/R283G) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Motojima, F, Yasukawa, K, Ohno, A, Asano, Y. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tailoring D-amino acid oxidase from the pid kidney to R-stereoselective amine oxidase and its use in the deracemization of 4-chlorobenzhydrylamine
To Be Published
|
|
5XAO
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala in complexes with sodium and chloride ions | | Descriptor: | ACETIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kamitori, S, Sode, K. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5ZJ9
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-3-carboxylic acid | | Descriptor: | 5-chloro thiophene-3-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
5ZJA
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-2-carboxylic acid | | Descriptor: | 5-chloro thiophene-2-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
6GG2
 
 | | The structure of FsqB from Aspergillus fumigatus, a flavoenzyme of the amine oxidase family | | Descriptor: | Amino acid oxidase fmpA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pavkov-Keller, T, Lahham, M, Macheroux, P, Gruber, K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Oxidative cyclization ofN-methyl-dopa by a fungal flavoenzyme of the amine oxidase family.
J. Biol. Chem., 293, 2018
|
|
6IWE
 
 | |
6J38
 
 | | Crystal structure of CmiS2 | | Descriptor: | FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2019-01-04 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6J39
 
 | | Crystal structure of CmiS2 with inhibitor | | Descriptor: | (3R)-3-[(carboxymethyl)sulfanyl]nonanoic acid, FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2019-01-04 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6KBP
 
 | | Crystal structure of human D-amino acid oxidase mutant (P219L) complexed with benzoate | | Descriptor: | ACETATE ION, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Rachadech, W, Kato, Y, Maita, N, Fukui, K. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human D-amino acid oxidase mutant (P219L) complexed with benzoate
To Be Published
|
|
6P9D
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6PLD
 
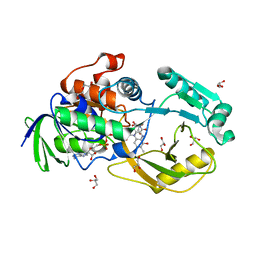 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6PXS
 
 | | Crystal structure of iminodiacetate oxidase (IdaA) from Chelativorans sp. BNC1 | | Descriptor: | FAD dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Jun, S.Y, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural and biochemical characterization of iminodiacetate oxidase from Chelativorans sp. BNC1.
Mol.Microbiol., 112, 2019
|
|
6RKF
 
 | | Structure of human DASPO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-aspartate oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Nardini, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.219 Å) | | Cite: | Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain.
Faseb J., 34, 2020
|
|
7CT4
 
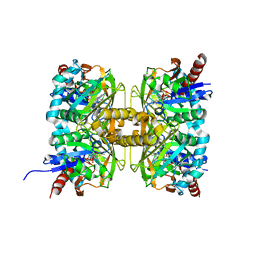 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7CYX
 
 | |
7EXS
 
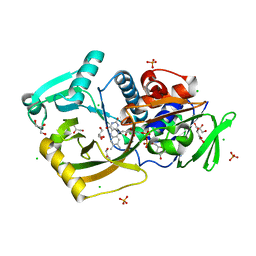 | | Thermomicrobium roseum sarcosine oxidase mutant - S320R | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xin, Y, Shen, C, Tang, M.W, Shi, Y, Guo, Z.T, Gu, Z.H, Shao, J, Zhang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Recreating the natural evolutionary trend in key microdomains provides an effective strategy for engineering of a thermomicrobial N-demethylase.
J.Biol.Chem., 298, 2022
|
|
7FGP
 
 | |
7LO1
 
 | |
7RDF
 
 | | Crystal structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F co-crystallized in the presence of D-arginine | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, A, Weber, I.T, Gadda, G. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery of a new flavin N5-adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase.
Arch.Biochem.Biophys., 715, 2021
|
|
7U9S
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | 5-{2-[4-(trifluoromethyl)phenyl]ethyl}-1,4-dihydropyrazine-2,3-dione, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7U9U
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | (3R)-3-(5,6-dioxo-1,4,5,6-tetrahydropyrazin-2-yl)-2,3-dihydro-1,4-benzoxathiine-7-carbonitrile, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7XQA
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry
Nat Commun, 14, 2023
|
|
7XX0
 
 | | C281S glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXC
 
 | | Orf1-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
