3FA0
 
 | |
3FAD
 
 | |
3FI5
 
 | |
3F9L
 
 | |
3G3X
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 100 K | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | 登録日 | 2009-02-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3V
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | 登録日 | 2009-02-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | 登録日 | 2009-02-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3GUP
 
 | |
3GUN
 
 | |
3GUJ
 
 | |
3GUO
 
 | |
3GUM
 
 | |
3GUI
 
 | |
3GUL
 
 | |
3GUK
 
 | |
3HDF
 
 | |
3HH3
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1,2-dihydro-1,2-azaborine | | 分子名称: | 1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2009-05-14 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH5
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1-ethyl-2-hydro-1,2-azaborine | | 分子名称: | 1-ethyl-1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2009-05-14 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH4
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - Benzene as control | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, BENZENE, Lysozyme, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2009-05-14 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH6
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity -ethylbenzene as control | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, PHENYLETHANE, ... | | 著者 | Liu, L, Matthews, B.W. | | 登録日 | 2009-05-14 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HDE
 
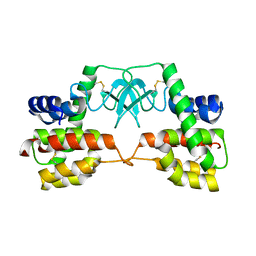 | | Crystal structure of full-length endolysin R21 from phage 21 | | 分子名称: | Lysozyme | | 著者 | Sun, Q, Arockiasamy, A, McKee, E, Caronna, E, Sacchettini, J.C. | | 登録日 | 2009-05-07 | | 公開日 | 2009-11-03 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Regulation of a muralytic enzyme by dynamic membrane topology.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HT8
 
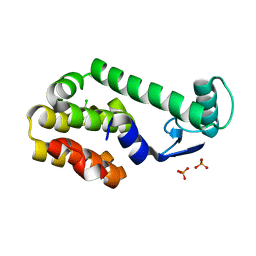 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | 分子名称: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
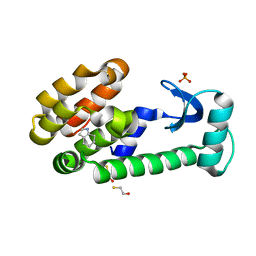 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU8
 
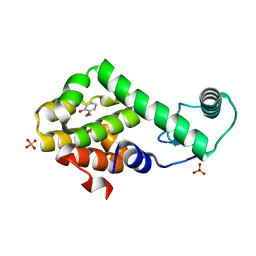 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
