5G5K
 
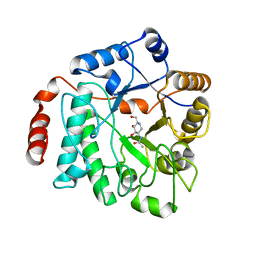 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G1M
 
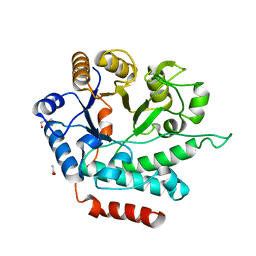 | | Crystal structure of NagZ from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, BETA-HEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5IOB
 
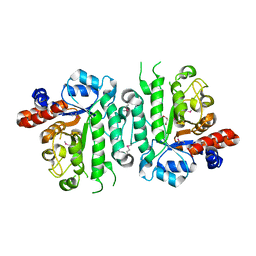 | | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase-related glycosidases, CHLORIDE ION, ... | | Authors: | Chang, C, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum
To Be Published
|
|
4HZM
 
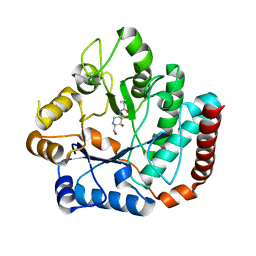 | | Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-hexosaminidase, N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Development of Selective Inhibitors of NagZ: Increased Susceptibility of Gram-Negative Bacteria to beta-Lactams.
Chembiochem, 14, 2013
|
|
4MSS
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
6JTI
 
 | |
6JTL
 
 | |
6JTJ
 
 | |
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
6GFV
 
 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
6JTK
 
 | | Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine | | Descriptor: | 2,2,2-tris(fluoranyl)-N-[(2R,3R,4R,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]ethanamide, Beta-hexosaminidase | | Authors: | Chen, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine
To Be Published
|
|
8C7F
 
 | |
6SZ6
 
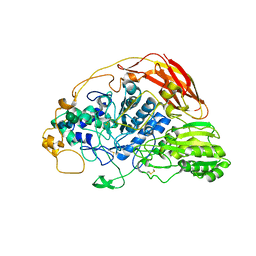 | | Chaetomium thermophilum beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Mohsin, I, Poudel, N, Papageorgiou, A.C. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Crystal Structure of a GH3 beta-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 20, 2019
|
|
1LQ2
 
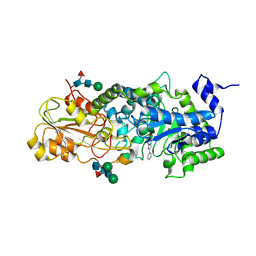 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
7PJJ
 
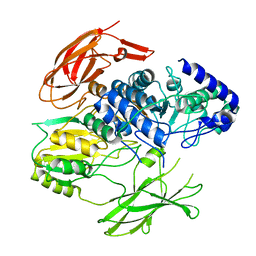 | | Structure of the Family-3 Glycosyl Hydrolase BcpE2 from Streptomyces scabies | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Jadot, C, Herman, R, Deflandre, B, Rigali, S, Kerff, F. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Structure and Function of BcpE2, the Most Promiscuous GH3-Family Glucose Scavenging Beta-Glucosidase.
Mbio, 13, 2022
|
|
7MS2
 
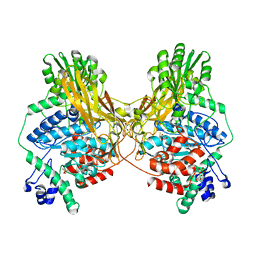 | |
8GYY
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose, 120 seconds | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, L.Y. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
3LK6
 
 | |
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase that is capable of degrading sesaminol triglucoside to produce sesaminol
To be published
|
|
3NVD
 
 | | Structure of YBBD in complex with pugnac | | Descriptor: | ACETATE ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, SODIUM ION, ... | | Authors: | Diederichs, K. | | Deposit date: | 2010-07-08 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism
J.Biol.Chem., 285, 2010
|
|
1X39
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-3-(ANILINOMETHYL)-5,6,7,8-TETRAHYDRO-5-(HYDROXYMETHYL)-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
1X38
 
 | | crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
7EY1
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
