6N9U
 
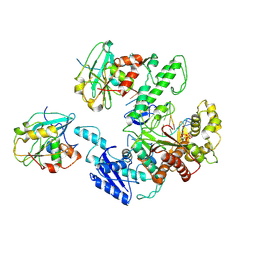 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) interacting with primase domains of two gp4 subunits bound to an RNA/DNA hybrid and dTTP (from LagS1) | | Descriptor: | DNA (44-MER), DNA primase/helicase, DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
5Z3N
 
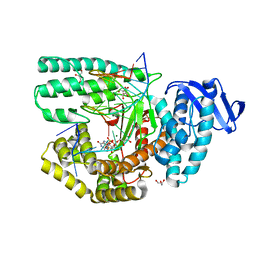 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base 5fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(5FC)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2018-01-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6N9X
 
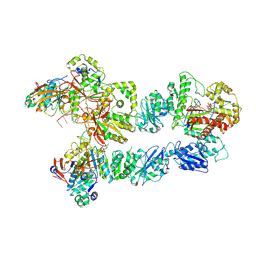 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS3) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N9W
 
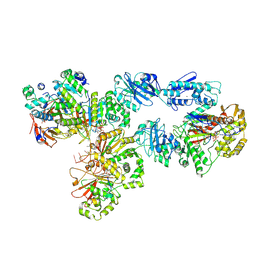 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS2) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
4XVM
 
 | |
4XVI
 
 | |
4XIU
 
 | |
4XVK
 
 | | Binary complex of human polymerase nu and DNA with the finger domain closed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*GP*AP*TP*CP*TP*GP*AP*CP*GP*CP*TP*AP*CP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*GP*TP*AP*GP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Lee, Y.-S, Yang, W. | | Deposit date: | 2015-01-27 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XVL
 
 | | Binary complex of human polymerase nu and DNA with the finger domain open | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*GP*AP*TP*CP*TP*GP*AP*CP*GP*CP*TP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*AP*GP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Lee, Y.-S, Yang, W. | | Deposit date: | 2015-01-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6VDE
 
 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
2KFZ
 
 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND ZINC ONLY | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KFN
 
 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-01 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KTQ
 
 | | OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-07-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
2KZZ
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZM
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
6FBI
 
 | | KlenTaq DNA polymerase in a closed, ternary complex with dGpNHpp bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBE
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the third primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*GP*GP*TP*GP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBF
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fourth primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*GP*GP*TP*GP*CP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*AP*(OH3)P*CP*GP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBC
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification at the 3'-terminus of the primer. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBD
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the second primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*AP*(OH3)P*C)-3'), DNA (5'-D(P*AP*AP*AP*CP*GP*GP*TP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBH
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the sixth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*GP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBG
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fifth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*CP*GP*GP*TP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3OJU
 
 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing c5 modified thymidies | | Descriptor: | 2'-deoxy-5-[(1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)ethynyl]uridine 5'-(tetrahydrogen triphosphate), DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OJS
 
 | | Snapshots of the large fragment of DNA polymerase I from Thermus Aquaticus processing C5 modified thymidines | | Descriptor: | 2'-deoxy-5-[9-(3-{[4-(diethylamino)-4-oxobutanoyl]amino}propyl)-18-ethyl-5,8,14,17-tetraoxo-4,9,13,18-tetraazaicos-1-yn-1-yl]uridine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PO5
 
 | | Structure of a mutant of the large fragment of DNA polymerase I from Thermus Auqaticus in complex with an abasic site and ddATP | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), DNA (5'-D(P*(3DR)P*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
