2N8S
 
 | |
2N8T
 
 | |
2N4W
 
 | |
2N4T
 
 | |
2N8U
 
 | |
2NNT
 
 | | General structural motifs of amyloid protofilaments | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Ferguson, N, Becker, J, Tidow, H, Tremmel, S, Sharpe, T.D, Krause, G, Flinders, J, Petrovich, M, Berriman, J, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | General structural motifs of amyloid protofilaments.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2MW9
 
 | |
2OEI
 
 | |
2OP7
 
 | | WW4 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Qin, H.N, Li, M.F, Pu, H, Sankaran, S, Ahmed, S, Song, J.X. | | Deposit date: | 2007-01-27 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the forth WW domain of WWP1
To be Published
|
|
3WH0
 
 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3KAF
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(1-benzothiophen-2-ylcarbonyl)-D-alanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7SA5
 
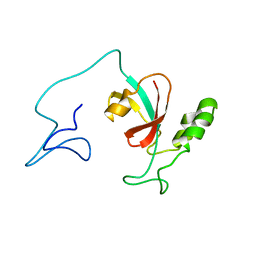 | | Two-state solution NMR structure of Apo Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Born, A, Vogeli, B. | | Deposit date: | 2021-09-22 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
7SUQ
 
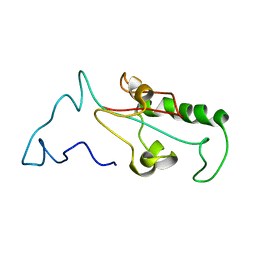 | |
7SUR
 
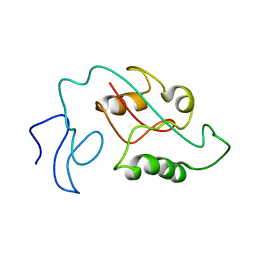 | |
1NMV
 
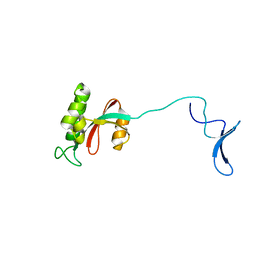 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
3TC5
 
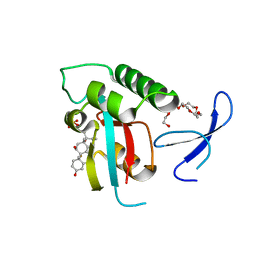 | | Selective targeting of disease-relevant protein binding domains by O-phosphorylated natural product derivatives | | Descriptor: | (11alpha,16alpha)-9-fluoro-11,17-dihydroxy-16-methyl-3,20-dioxopregna-1,4-dien-21-yl dihydrogen phosphate, HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Graeber, M, Janczyk, W, Sperl, B, Elumalai, N, Kozany, C, Hausch, F, Holak, T.A, Berg, T. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective targeting of disease-relevant protein binding domains by o-phosphorylated natural product derivatives.
Acs Chem.Biol., 6, 2011
|
|
3TDB
 
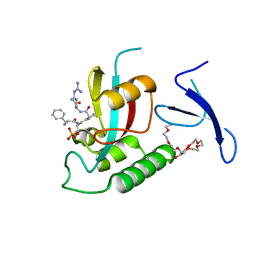 | | Human Pin1 bound to trans peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N-[(1E,2R)-1-[(2R)-2-{[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]carbamoyl}cyclopentylidene]-3-(phosphonooxy)propan-2-yl]-L-phenylalaninamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-10 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
3TCZ
 
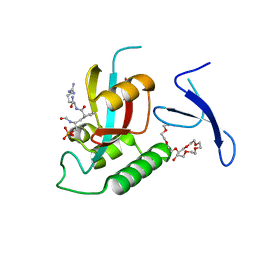 | | Human Pin1 bound to cis peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N~2~-({(1R,2Z)-2-[(2R)-2-(formylamino)-3-(phosphonooxy)propylidene]cyclopentyl}carbonyl)-L-argininamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
1EG3
 
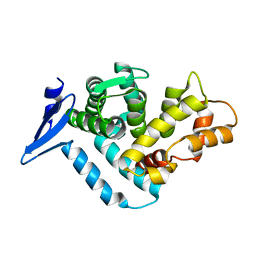 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EG4
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | BETA-DYSTROGLYCAN, DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1PIN
 
 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
5XMC
 
 | |
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
