5DOM
 
 | | Crystal structure, maturation and flocculating properties of a 2S albumin from Moringa oleifera seeds | | Descriptor: | 1,2-ETHANEDIOL, 2S albumin, ACETATE ION | | Authors: | Ullah, A, Murakami, M.T, Arni, R.K. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of mature 2S albumin from Moringa oleifera seeds.
Biochem.Biophys.Res.Commun., 468, 2015
|
|
3GSH
 
 | | Three-dimensional structure of a post translational modified barley LTP1 | | Descriptor: | (12E)-10-oxooctadec-12-enoic acid, Non-specific lipid-transfer protein 1, SODIUM ION, ... | | Authors: | Lascombe, M.B, Prange, T, Bakan, B, Marion, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of oxylipin-conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid-binding proteins.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
6S3F
 
 | | Moringa seed protein Mo-CBP3-4 | | Descriptor: | 2S albumin, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Moulin, M, Mossou, E, Mitchell, E.P, Haertlein, M, Forsyth, V.T, Rennie, A.R. | | Deposit date: | 2019-06-25 | | Release date: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Towards a molecular understanding of the water purification properties of Moringa seed proteins.
J Colloid Interface Sci, 554, 2019
|
|
4XUW
 
 | | Structure of the hazelnut allergen, Cor a 8 | | Descriptor: | Non-specific lipid-transfer protein, PHOSPHATE ION | | Authors: | Offermann, L.R, Perdue, M.L, Bublin, M, Pfeifer, S, Dubiela, P, Hoffmann-Sommergruber, K, Chruszcz, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.
J.Agric.Food Chem., 63, 2015
|
|
1W2Q
 
 | |
7W9A
 
 | |
6FRR
 
 | | Structural and immunological properties of the allergen Art v 3 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Brandstetter, H, Soh, W.T, Magler, I. | | Deposit date: | 2018-02-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Boiling down the cysteine-stabilized LTP fold - loss of structural and immunological integrity of allergenic Art v 3 and Pru p 3 as a consequence of irreversible lanthionine formation.
Mol.Immunol., 116, 2019
|
|
6VJ0
 
 | | Crystal structure of a chitin-binding protein from Moringa oleifera seeds (Mo-CBP4) | | Descriptor: | ACETATE ION, CHLORIDE ION, Chitin-binding protein Mo-CBP4 | | Authors: | Bezerra, E.H.S, Lopes, T.D.P, da Silva, F.M.S, Costa, H.P.S, Freire, V.N, Rocha, B.A.M, Sousa, D.O.B. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a lectin from Moringa oleifera seeds with imflammatory activities
To Be Published
|
|
5LQV
 
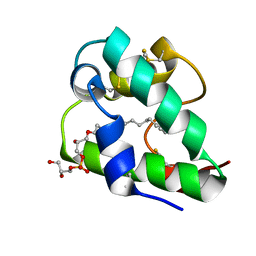 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
8DB4
 
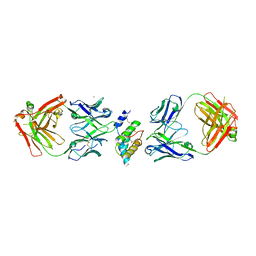 | |
8G4P
 
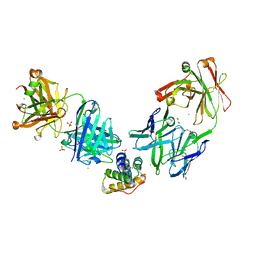 | | Crystal structure of the peanut allergen Ara h 2 bound by two neutralizing antibodies 13T1 and 13T5 | | Descriptor: | 1,2-ETHANEDIOL, 13T1 Fab light chain, 13T5 Fab heavy chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design of an Ara h 2 hypoallergen from conformational epitopes.
Clin Exp Allergy, 54, 2024
|
|
1L6H
 
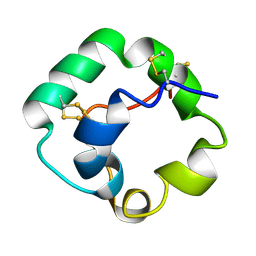 | |
1LIP
 
 | |
7KSB
 
 | | Crystal structure on Act c 10.0101 | | Descriptor: | Non-specific lipid-transfer protein 1, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Giangrieco, I, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
7KSC
 
 | | Crystal structure of Pun g 1.0101 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Tuppo, L, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
5TVI
 
 | |
5U87
 
 | | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin | | Descriptor: | Preproalbumin PawS1 | | Authors: | Franke, B, James, A.M, Mobli, M, Colgrave, M.L, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin
to be published
|
|
1HYP
 
 | |
1JTB
 
 | | LIPID TRANSFER PROTEIN COMPLEXED WITH PALMITOYL COENZYME A, NMR, 16 STRUCTURES | | Descriptor: | COENZYME A, LIPID TRANSFER PROTEIN, PALMITIC ACID | | Authors: | Lerche, M.H, Kragelund, B.B, Bech, L.M, Poulsen, F.M. | | Deposit date: | 1996-12-03 | | Release date: | 1997-07-07 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands.
Structure, 5, 1997
|
|
2DS2
 
 | | Crystal structure of mabinlin II | | Descriptor: | ACETIC ACID, Sweet protein mabinlin-2 chain A, Sweet protein mabinlin-2 chain B | | Authors: | Li, D.F, Zhu, D.Y, Wang, D.C. | | Deposit date: | 2006-06-19 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.
J.Struct.Biol., 162, 2008
|
|
1HSS
 
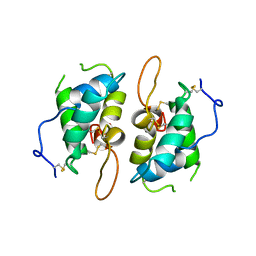 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
2B5S
 
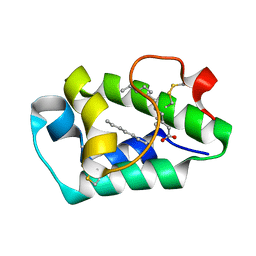 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2ALG
 
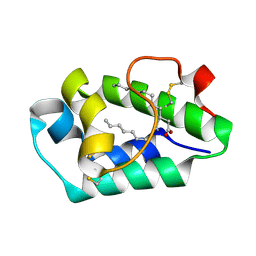 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2LVF
 
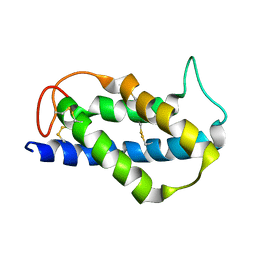 | | Solution structure of the Brazil Nut 2S albumin Ber e 1 | | Descriptor: | 2S albumin | | Authors: | Rundqvist, L, Tengel, T, Zdunek, J, Schleucher, J, Alcocer, M.J, Larsson, G. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure, copper binding and backbone dynamics of recombinant Ber e 1-the major allergen from Brazil nut.
Plos One, 7, 2012
|
|
2N2Z
 
 | | NMR spatial structure of nonspecific lipid transfer protein from the dill Anethum graveolens L. | | Descriptor: | Non-specific lipid-transfer protein | | Authors: | Mineev, K.S, Melnikova, D.N, Finkina, E.I, Arseniev, A.S, Ovchinnikova, T.V. | | Deposit date: | 2015-05-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the dill Anethum graveolens L.: isolation, structure, heterologous expression, and functional characteristics.
J.Pept.Sci., 22, 2016
|
|
