7OO3
 
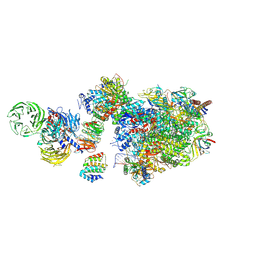 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | 分子名称: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-26 | | 公開日 | 2021-10-06 | | 最終更新日 | 2021-10-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPD
 
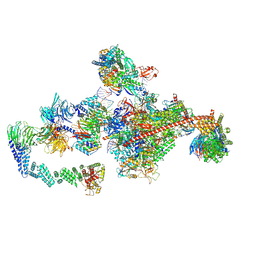 | |
7OOB
 
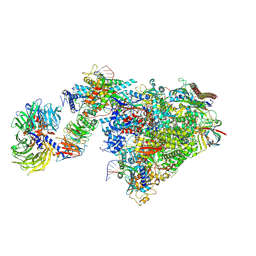 | |
7OPC
 
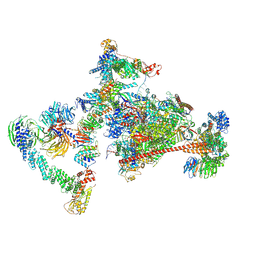 | |
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | 分子名称: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | 登録日 | 2023-08-18 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6LTH
 
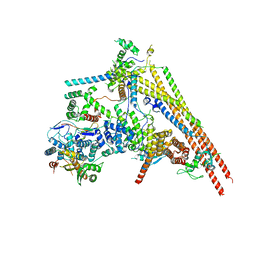 | | Structure of human BAF Base module | | 分子名称: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | 著者 | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | 登録日 | 2020-01-22 | | 公開日 | 2020-02-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
7Z8S
 
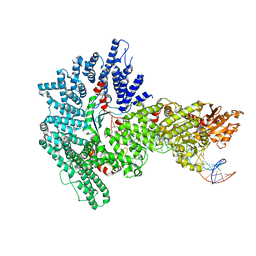 | | Mot1:TBP:DNA - post hydrolysis state | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-18 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
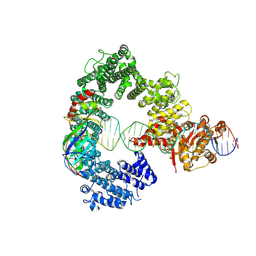 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-16 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
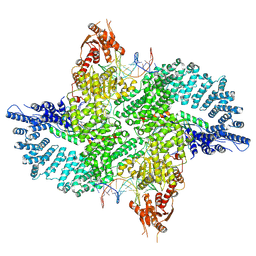 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-23 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZKE
 
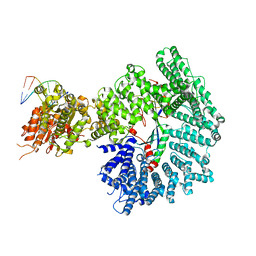 | | Mot1:TBP:DNA - pre-hydrolysis state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PWF
 
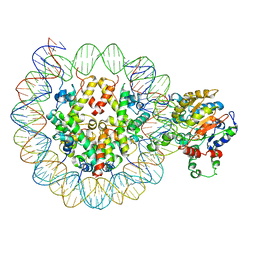 | |
7OTQ
 
 | | Cryo-EM structure of ALC1/CHD1L bound to a PARylated nucleosome | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Deindl, S. | | 登録日 | 2021-06-10 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome
Elife, 10, 2021
|
|
8EUF
 
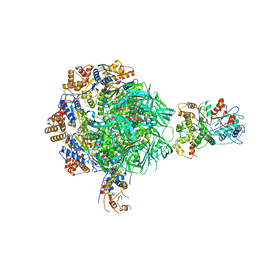 | | Class2 of the INO80-Nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | 登録日 | 2022-10-18 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8HE5
 
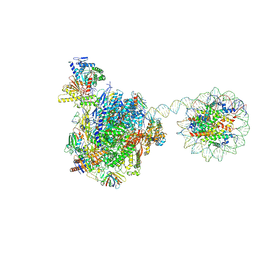 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | 分子名称: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-11-07 | | 公開日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (6.95 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
5O9G
 
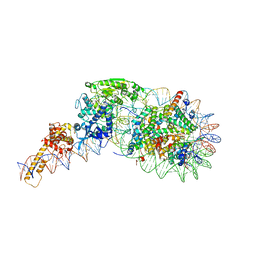 | | Structure of nucleosome-Chd1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | 著者 | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | 登録日 | 2017-06-19 | | 公開日 | 2017-10-11 | | 最終更新日 | 2017-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
6RYR
 
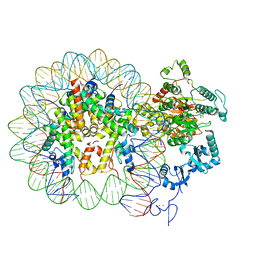 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | 分子名称: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | 著者 | Farnung, L, Ochmann, M, Cramer, P. | | 登録日 | 2019-06-11 | | 公開日 | 2020-07-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6RYU
 
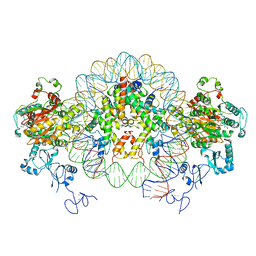 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | 分子名称: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | 著者 | Farnung, L, Ochmann, M, Cramer, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-07-15 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
8B0A
 
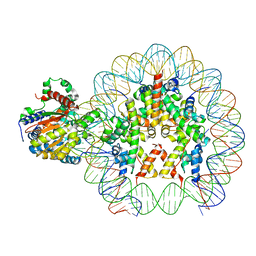 | | Cryo-EM structure of ALC1 bound to an asymmetric, site-specifically PARylated nucleosome | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Deindl, S. | | 登録日 | 2022-09-07 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1.
Nat Commun, 15, 2024
|
|
7UX9
 
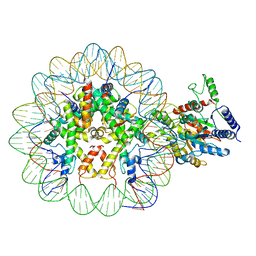 | | Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA) | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | 著者 | Ipsaro, J.J, Adams, D.W, Joshua-Tor, L. | | 登録日 | 2022-05-05 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Cell, 186, 2023
|
|
7VDT
 
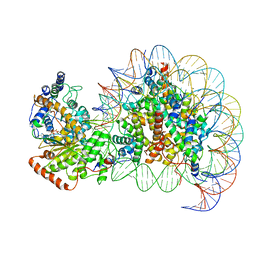 | | The motor-nucleosome module of human chromatin remodeling PBAF-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (207-MER), ... | | 著者 | Chen, Z.C, Chen, K.J, Yuan, J.J. | | 登録日 | 2021-09-07 | | 公開日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
7VDV
 
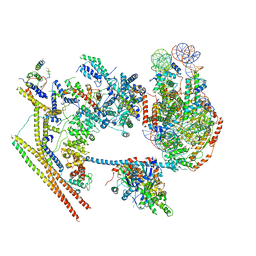 | | The overall structure of human chromatin remodeling PBAF-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2,AT-rich interactive domain-containing protein 2, Actin, ... | | 著者 | Chen, Z.C, Chen, K.J, Yuan, J.J. | | 登録日 | 2021-09-07 | | 公開日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
7R76
 
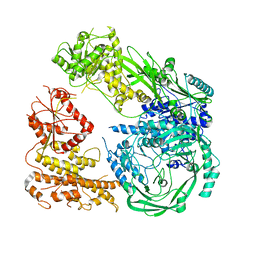 | | Cryo-EM structure of DNMT5 in apo state | | 分子名称: | DNA repair protein Rad8, ZINC ION | | 著者 | Wang, J, Patel, D.J. | | 登録日 | 2021-06-24 | | 公開日 | 2022-02-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
7R77
 
 | |
7R78
 
 | |
6IRO
 
 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | 著者 | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | 登録日 | 2018-11-13 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
