6UEZ
 
 | |
6UDL
 
 | |
6UDM
 
 | |
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
6U30
 
 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
6U31
 
 | | The crystal structure of 4-(1H-imidazol-1-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(1H-imidazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
6Q2T
 
 | | Human sterol 14a-demethylase (CYP51) in complex with the functionally irreversible inhibitor (R)-N-(1-(3-chloro-4'-fluoro-[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Lanosterol 14-alpha demethylase, N-[(1R)-1-(3-chloro-4'-fluoro[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, ... | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Validation of Human Sterol 14 alpha-Demethylase (CYP51) Druggability: Structure-Guided Design, Synthesis, and Evaluation of Stoichiometric, Functionally Irreversible Inhibitors.
J.Med.Chem., 62, 2019
|
|
6Q2C
 
 | | Domain-swapped dimer of Acanthamoeba castellanii CYP51 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-08-07 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
6PRR
 
 | | The crystal structure of 3-methylaminobenzoate-bound CYP199A4 | | Descriptor: | 3-(methylamino)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-11 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6PRS
 
 | | The crystal structure of 3-ethoxybenzoate-bound CYP199A4 | | Descriptor: | 3-ethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-11 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6PQS
 
 | | The crystal structure of 4-methylbenzoate-bound CYP199A4 | | Descriptor: | 4-METHYLBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6PQW
 
 | | The crystal structure of 3-methylbenzoate-bound CYP199A4 | | Descriptor: | 3-methylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6KFW
 
 | |
6PQD
 
 | | The crystal structure of 3-methylthiobenzoate-bound CYP199A4 | | Descriptor: | 3-(methylsulfanyl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6PQ6
 
 | | The crystal structure of 3-methoxybenzoate-bound CYP199A4 | | Descriptor: | 3-methoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bell, S.G, Bruning, J.B. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
6KBH
 
 | | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase | | Descriptor: | Cytochrome P450 monooxygenase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gong, R, Wu, L.J, Zhang, Y, Liu, Z, Dou, S, Zhang, R.G, Xu, J.H, Tang, C, Zhou, J.H. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase
To Be Published
|
|
6K9S
 
 | | Structure of the Carbonylruthenium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CARBON MONOXIDE, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6K58
 
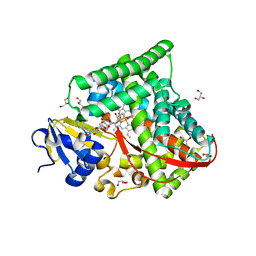 | | Structure of the CYP102A1 Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6K3Q
 
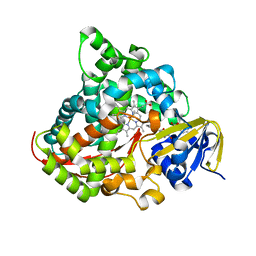 | | Crystal Structure of P450BM3 with N-(3-cyclohexylpropanoyl)-L-prolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-(3-cyclohexylpropanoyl)pyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6RQ3
 
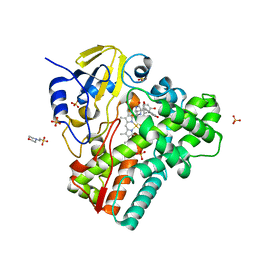 | | CYP121 in complex with 2,6-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(2,6-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ8
 
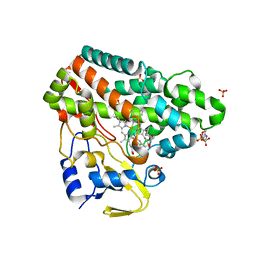 | | CYP121 in complex with 3-iodo dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-iodanyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ0
 
 | | CYP121 in complex with 3-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQD
 
 | | CYP121 in complex with 3-chloro dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3-chloranyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ1
 
 | | CYP121 in complex with 2-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(2-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQB
 
 | | CYP121 in complex with 3-bromo dicyclotyrosine | | Descriptor: | 3-bromo dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
