8W2L
 
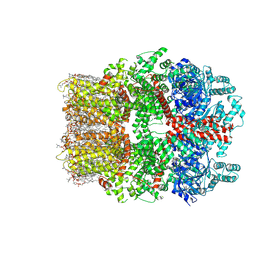 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | Authors: | Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
8T6L
 
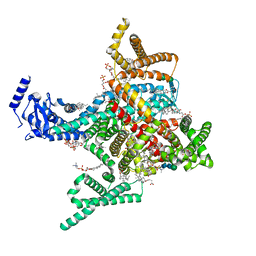 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
4EN1
 
 | |
6AA6
 
 | |
6EFR
 
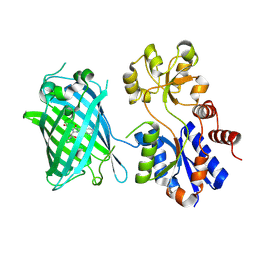 | | Crystal Structure of iNicSnFR 1.0 | | Descriptor: | iNicSnFR 1.0, a genetically encoded nicotine biosensor,Green fluorescent protein | | Authors: | Shivange, A.V, Borden, P.M. | | Deposit date: | 2018-08-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nicotinic Drugs in the Endoplasmic Reticulum: Beginning the Inside-out Pathway of Addiction and Therapy
J.Gen.Physiol., 2019
|
|
8A6O
 
 | |
8A6P
 
 | |
8A6R
 
 | |
8A6G
 
 | |
8A83
 
 | |
8A6N
 
 | |
8A6Q
 
 | |
8A6S
 
 | |
8A7V
 
 | |
8AM4
 
 | | Cl-rsEGFP2 Long Wavelength Structure | | Descriptor: | Green fluorescent protein | | Authors: | Orr, C.M, Fadini, A, van Thor, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Serial Femtosecond Crystallography Reveals that Photoactivation in a Fluorescent Protein Proceeds via the Hula Twist Mechanism.
J.Am.Chem.Soc., 2023
|
|
8BVG
 
 | |
8BXP
 
 | |
8C1X
 
 | |
4XBI
 
 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
6L27
 
 | | X-ray crystal structure of the mutant green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
5LEL
 
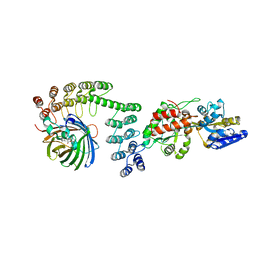 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEM
 
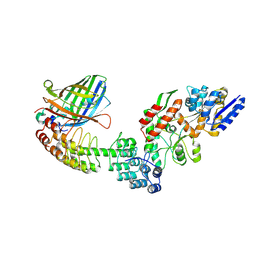 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
6FLL
 
 | |
1EML
 
 | |
1EMK
 
 | |
