4AYP
 
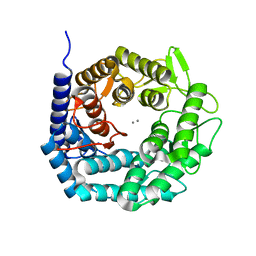 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside | | 分子名称: | CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, SODIUM ION, ... | | 著者 | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | 登録日 | 2012-06-21 | | 公開日 | 2013-01-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYO
 
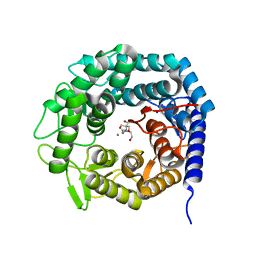 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | 著者 | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | 登録日 | 2012-06-21 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7UCP
 
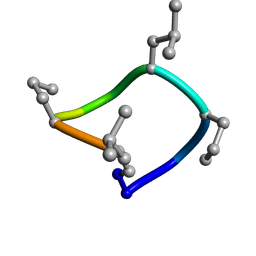 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
6JGI
 
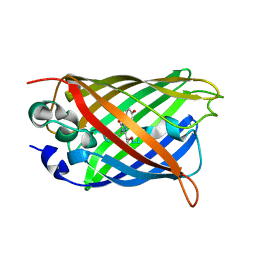 | | Crystal structure of the S65T/F99S/M153T/V163A variant of GFP at 0.85 A | | 分子名称: | Green fluorescent protein | | 著者 | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
3O4P
 
 | | DFPase at 0.85 Angstrom resolution (H atoms included) | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | 著者 | Liebschner, D, Elias, M, Koepke, J, Lecomte, C, Guillot, B, Jelsch, C, Chabriere, E. | | 登録日 | 2010-07-27 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Hydrogen atoms in protein structures: high-resolution X-ray diffraction structure of the DFPase.
BMC Res Notes, 6, 2013
|
|
6ETL
 
 | |
6ETK
 
 | |
6Q00
 
 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | 分子名称: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | 著者 | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | 登録日 | 2019-08-01 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-06-24 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
1PJX
 
 | | 0.85 ANGSTROM STRUCTURE OF SQUID GANGLION DFPASE | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | 著者 | Koepke, J, Rueterjans, H, Luecke, C, Fritzsch, G. | | 登録日 | 2003-06-04 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Statistical analysis of crystallographic data obtained from squid ganglion DFPase at 0.85 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4HIF
 
 | | Ultrahigh-resolution crystal structure of Z-DNA in complex with Zn2+ ions | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE (FULLY PROTONATED FORM), ... | | 著者 | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | 登録日 | 2012-10-11 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6AIR
 
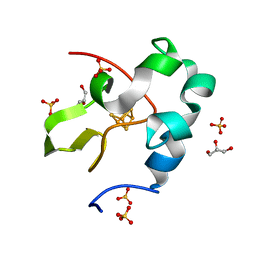 | | High resolution structure of perdeuterated high-potential iron-sulfur protein | | 分子名称: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | 著者 | Hanazono, Y, Takeda, K, Miki, K. | | 登録日 | 2018-08-24 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
7RVE
 
 | |
3U7T
 
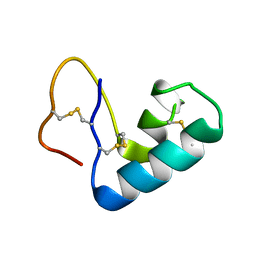 | |
6AIQ
 
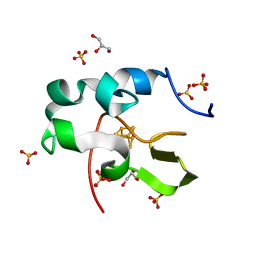 | | High resolution structure of recombinant high-potential iron-sulfur protein | | 分子名称: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | 著者 | Hanazono, Y, Takeda, K, Miki, K. | | 登録日 | 2018-08-24 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
7P4R
 
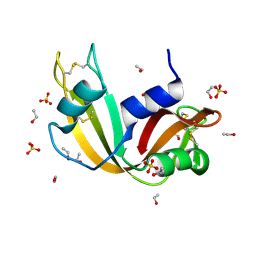 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | 分子名称: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | 著者 | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | 登録日 | 2021-07-12 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
2OL9
 
 | |
4I8K
 
 | | Bovine trypsin at 0.85 resolution | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | 登録日 | 2012-12-03 | | 公開日 | 2012-12-19 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PQ5
 
 | | Trypsin at pH 5, 0.85 A | | 分子名称: | ARGININE, SULFATE ION, Trypsin | | 著者 | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | 登録日 | 2003-06-18 | | 公開日 | 2003-11-11 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
5VLE
 
 | |
4D5M
 
 | | Gonadotropin-releasing hormone agonist | | 分子名称: | PHOSPHATE ION, TRIPTORELIN | | 著者 | Legrand, P, Le Du, M.-H, Valery, C, Deville-Foillard, S, Paternostre, M, Artzner, F. | | 登録日 | 2014-11-05 | | 公開日 | 2015-08-12 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Atomic View of the Histidine Environment Stabilizing Higher- Ph Conformations of Ph-Dependent Proteins.
Nat.Commun., 6, 2015
|
|
7KQO
 
 | |
2PF8
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is equal to concentration of IDD594. | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | 登録日 | 2007-04-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
5EMB
 
 | |
4IGS
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | 分子名称: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2012-12-18 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | 分子名称: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | 著者 | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | 登録日 | 2013-12-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
