6AHH
 
 | | Crystal Structure of HEWL in complex with Phenylethyl alcohol (in the aroma form) after 5 hours under fibrillation conditions | | Descriptor: | 2-PHENYL-ETHANOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Seraj, Z, Seyedarabi, A. | | Deposit date: | 2018-08-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cinnamaldehyde and Phenyl Ethyl Alcohol promote the entrapment of intermediate species of HEWL, as revealed by structural, kinetics and thermal stability studies.
Sci Rep, 9, 2019
|
|
6PXU
 
 | | Crystal structure of human GalNAc-T12 bound to a diglycosylated peptide, Mn2+, and UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GAGATGAGAGYYITPRTGAGA, ... | | Authors: | Samara, N.L, Fernandez, A.J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | The structure of the colorectal cancer-associated enzyme GalNAc-T12 reveals how nonconserved residues dictate its function.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9GTF
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 57256190 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-17 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
5TC0
 
 | |
7AYA
 
 | | Crystal structure of CK2 bound by compound 9 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
5TD2
 
 | |
5K6Q
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase catalytic subunit | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Wang, J.-G, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Crystal structure of plant acetohydroxyacid synthase, the target for several commercial herbicides.
FEBS J., 284, 2017
|
|
6Q39
 
 | | Complex of Arginase 2 with Example 49 | | Descriptor: | 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-1-[[(2~{S})-piperidin-2-yl]methyl]pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, mitochondrial, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6HB5
 
 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-cytidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine--tRNA ligase, ... | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
6EDA
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-09 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
7DLR
 
 | | Mycobacterium tuberculosis enolase mutant - E163A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ahmad, M, Biswal, B.K. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
9GQ2
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,1)-(E) | | Descriptor: | (2~{S},9~{S},13~{E})-2-cyclohexyl-18,21-dimethoxy-11,16-dioxa-4-azatricyclo[15.2.2.0^{4,9}]henicosa-1(19),13,17,20-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8AB8
 
 | | Complex III2, b-position, with decylubiquinone and ascorbate-reduced | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
7B3X
 
 | | Notum complex with ARUK3003748 | | Descriptor: | 6-(m-tolylthio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
6TOH
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Shetty, K, Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6JZ0
 
 | | Crystal structure of EGFR kinase domain in complex with compound 78 | | Descriptor: | E-4-(dimethylamino)-N-[3-[4-[[(1S)-2-oxidanyl-1-phenyl-ethyl]amino]-6-phenyl-furo[2,3-d]pyrimidin-5-yl]phenyl]but-2-enamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a Furanopyrimidine-Based Epidermal Growth Factor Receptor Inhibitor (DBPR112) as a Clinical Candidate for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 62, 2019
|
|
8AB7
 
 | | Complex III2 from Yarrowia lipolytica, atovaquone and antimycin A bound | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABL
 
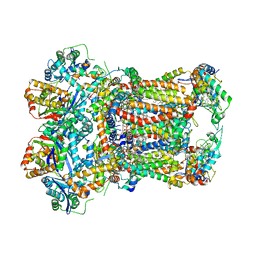 | | Complex III2 from Yarrowia lipolytica, with decylubiquinol and antimycin A, consensus refinement | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
6JZU
 
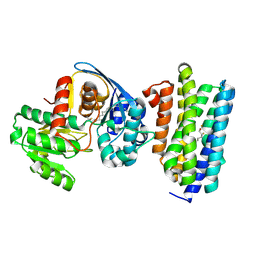 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
5TDX
 
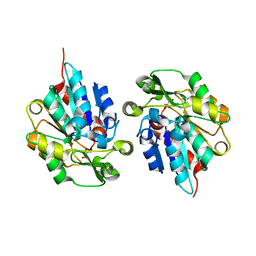 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
8ABK
 
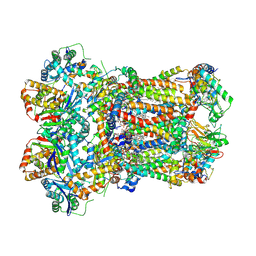 | | Complex III2 from Yarrowia lipolytica, decylubiquinol bound, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
9GGO
 
 | | Strand-swapped dimer of engineered copper binding SH3-like protein | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SH3-like protein | | Authors: | Schwan, M, Kopp, J, Sinning, I. | | Deposit date: | 2024-08-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strand-Swapped SH3 Domain Dimer with Superoxide Dismutase Activity.
Acs Cent.Sci., 11, 2025
|
|
9GPY
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12g/j | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6E91
 
 | | CA IX mimic Complexed with Steroidal Sulfamate Compound STX 2484 | | Descriptor: | 7-methoxy-6-(sulfamoyloxy)-2-[(3,4,5-trimethoxyphenyl)methyl]isoquinolin-2-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, Mckenna, R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3,17 beta-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene and Nonsteroidal Sulfamate Derivatives Inhibit Carbonic Anhydrase IX: Structure-Activity Optimization for Isoform Selectivity.
J. Med. Chem., 62, 2019
|
|
6HLX
 
 | | Structure of the PBP AgaA in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
