2F2A
 
 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Gln | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, GLUTAMINE, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
3A2Z
 
 | | E. coli Gsp amidase Cys59 sulfenic acid | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase | | Authors: | Pai, C.-H, Ko, T.-P, Chiang, B.-Y, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-06-05 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation
J.Biol.Chem., 285, 2010
|
|
2DQN
 
 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Asn | | Descriptor: | ASPARAGINE, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-05-29 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
3A2Y
 
 | | E. coli Gsp amidase C59A complexed with Gsp | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE | | Authors: | Pai, C.-H, Ko, T.-P, Chiang, B.-Y, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
2E18
 
 | |
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
2DZD
 
 | | Crystal structure of the biotin carboxylase domain of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Sueda, S, Islam, M.N, Kondo, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3A30
 
 | | E. coli Gsp amidase C59 acetate modification | | Descriptor: | ACETATE ION, Bifunctional glutathionylspermidine synthetase/amidase | | Authors: | Pai, C.-H, Ko, T.-P, Chiang, B.-Y, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-06-05 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation
J.Biol.Chem., 285, 2010
|
|
1Z56
 
 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
3UQ8
 
 | |
3J82
 
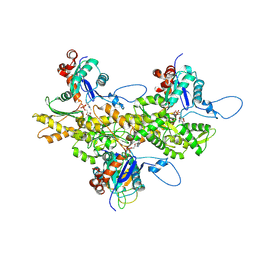 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
2EF7
 
 | | Crystal structure of ST2348, a hypothetical protein with CBS domains from Sulfolobus tokodaii strain7 | | Descriptor: | Hypothetical protein ST2348 | | Authors: | Agari, Y, Karthe, P, Kumarevel, T, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-21 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ST2348, a CBS domain protein, from hyperthermophilic archaeon Sulfolobus tokodaii
Biochem.Biophys.Res.Commun., 375, 2008
|
|
1GSA
 
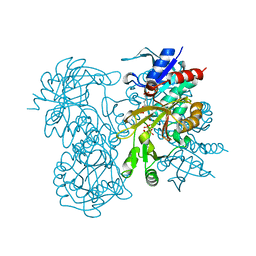 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
1HRU
 
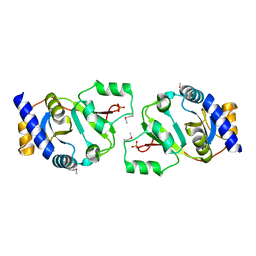 | | THE STRUCTURE OF THE YRDC GENE PRODUCT FROM E.COLI | | Descriptor: | PHOSPHATE ION, YRDC GENE PRODUCT | | Authors: | Teplova, M, Tereshko, V, Sanishvili, R, Joachimiak, A, Bushueva, T, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
Protein Sci., 9, 2000
|
|
1GTD
 
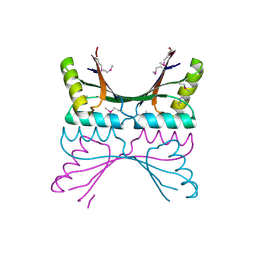 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID TT50) STRUCTURE OF MTH169, THE PURS SUBUNIT OF FGAM SYNTHETASE | | Descriptor: | MTH169 | | Authors: | Batra, R, Christendat, D, Saxild, H.H, Arrowsmith, C, Tong, L. | | Deposit date: | 2002-01-14 | | Release date: | 2002-12-12 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal Structure of Mth169, a Crucial Component of Phosphoribosylformylglycinamidine Synthetase
Proteins: Struct.,Funct., Genet., 49, 2002
|
|
1IMO
 
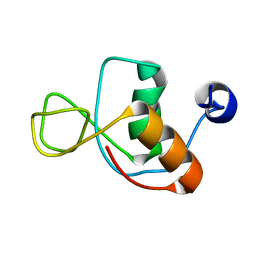 | |
1IK9
 
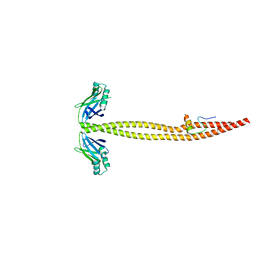 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
1IN1
 
 | |
1TYC
 
 | |
1TYD
 
 | |
1TYB
 
 | |
7YBU
 
 | | Human propionyl-coenzyme A carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Su, J.Y, Liu, D.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human propionyl-coenzyme A carboxylase
To Be Published
|
|
1UW0
 
 | |
7ZZ5
 
 | | Cryo-EM structure of "BC open" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, BICARBONATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYZ
 
 | | Cryo-EM structure of "CT oxa" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
