3K8D
 
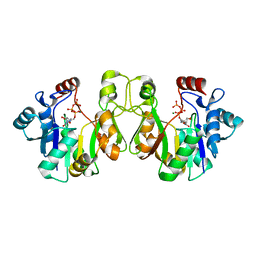 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
4WFE
 
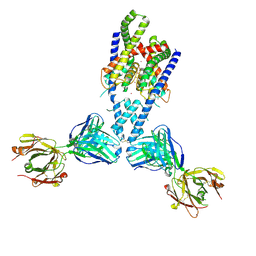 | | Human TRAAK K+ channel in a K+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
3SKM
 
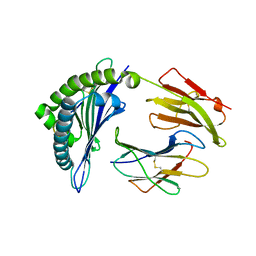 | | Crystal structure of the HLA-B8FLRGRAYVL, mutant G8V of the FLR peptide | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
4WH1
 
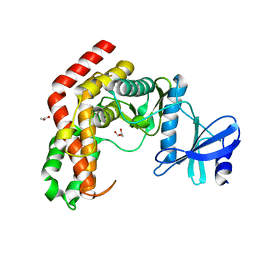 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
3SH5
 
 | |
3SL3
 
 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4WKJ
 
 | | Crystallographic Structure of a Dodecameric RNA-DNA Hybrid | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3') | | Authors: | Davis, R.R, Shaban, N.M, Perrino, F.W, Hollis, T. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of RNA-DNA duplex provides insight into conformational changes induced by RNase H binding.
Cell Cycle, 14, 2015
|
|
3SLL
 
 | |
4WIJ
 
 | | HUMAN SPLICING FACTOR, CONSTRUCT 1 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | lee, M, bond, c.s. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation.
Nucleic Acids Res., 43, 2015
|
|
3SHZ
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3JVG
 
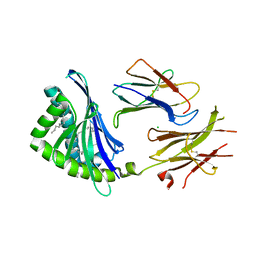 | | Crystal Structure of chicken CD1-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Dvir, H, Wang, J, Zajonc, D.M. | | Deposit date: | 2009-09-16 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for lipid-antigen recognition in avian immunity.
J.Immunol., 184, 2010
|
|
4WL1
 
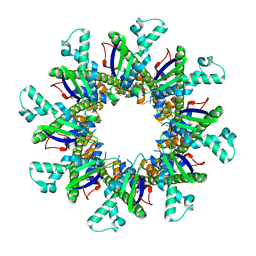 | |
3SNV
 
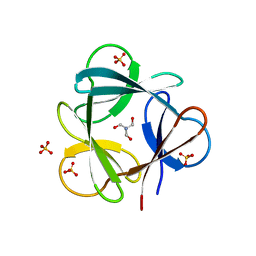 | |
3SO5
 
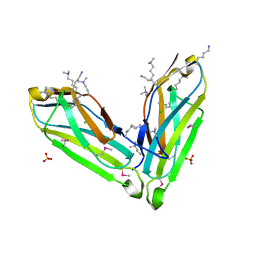 | |
4WO2
 
 | |
4WO7
 
 | |
3S1V
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-fructose 6-phosphate Schiff-base intermediate | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
4WPM
 
 | |
4WQ7
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - "as isolated" form | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
3SJ7
 
 | |
3S2E
 
 | | Crystal Structure of FurX NADH Complex 1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4WL3
 
 | |
3S3D
 
 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 480s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
3J77
 
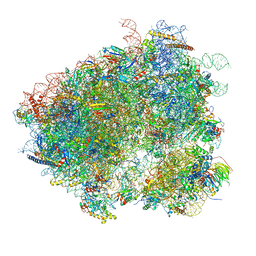 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3S3U
 
 | |
