1UZU
 
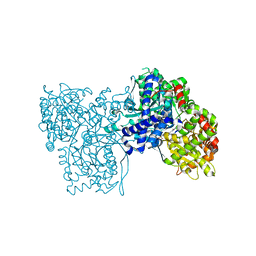 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
1UZV
 
 | | High affinity fucose binding of Pseudomonas aeruginosa lectin II: 1.0 A crystal structure of the complex | | Descriptor: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | Authors: | Mitchell, E, Sabin, C.D, Snajdrova, L, Budova, M, Perret, S, Gautier, C, Gilboa-Garber, N, Koca, J, Wimmerova, M, Imberty, A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin Pa-Iil: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches.
Proteins, 58, 2005
|
|
1UZW
 
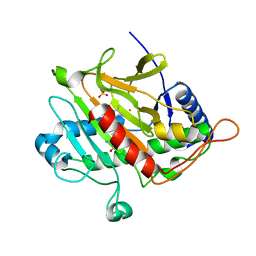 | | ISOPENICILLIN N SYNTHASE WITH L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE | | Descriptor: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Grummitt, A.R, Rutledge, P.J, Clifton, I.J, Baldwin, J.E. | | Deposit date: | 2004-03-17 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Active Site Mediated Elimination of Hydrogen Fluoride from a Fluorinated Substrate Analogue by Isopenicillin N Synthase
Biochem.J., 382, 2004
|
|
1UZX
 
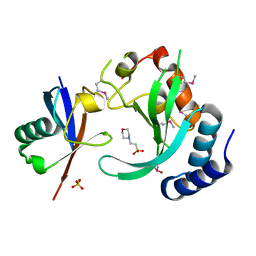 | | A complex of the Vps23 UEV with ubiquitin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UBIQUITIN, ... | | Authors: | Teo, H, Williams, R.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-03-30 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
J.Biol.Chem., 279, 2004
|
|
1UZY
 
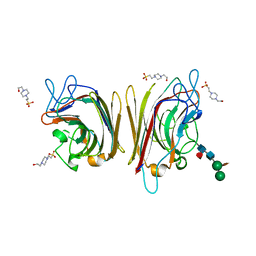 | | Erythrina crystagalli lectin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Lectin, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Erythrina cristagalli lectin with bound N-linked oligosaccharide and lactose.
Glycobiology, 14, 2004
|
|
1UZZ
 
 | | Erythrina cristagalli bound to N-linked oligosaccharide and lactose | | Descriptor: | CALCIUM ION, GLYCEROL, Lectin, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of Erythrina cristagalli lectin with bound N-linked oligosaccharide and lactose.
Glycobiology, 14, 2004
|
|
1V00
 
 | | Erythrina cristagalli lectin | | Descriptor: | CALCIUM ION, LECTIN (ECL), MANGANESE (II) ION, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-21 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Erythrina Cristagalli Lectin with Bound N-Linked Oligosaccharide and Lactose
Glycobiology, 14, 2004
|
|
1V02
 
 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | DHURRINASE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V03
 
 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | ACETONITRILE, DHURRINASE, PHENOL, ... | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V04
 
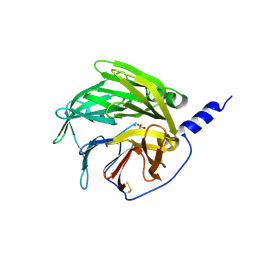 | | serum paraoxonase by directed evolution | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SERUM PARAOXONASE/ARYLESTERASE 1 | | Authors: | Harel, M, Aharoni, A, Gaidukov, L, Brumshtein, B, Khersonsky, O, Yagur, S, Meged, R, Dvir, H, Ravelli, R.B.G, McCarthy, A, Toker, L, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-23 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Evolution of the Serum Paraoxonase Family of Detoxifying and Anti-Atherosclerotic Enzymes
Nat.Struct.Mol.Biol., 11, 2004
|
|
1V05
 
 | |
1V06
 
 | |
1V07
 
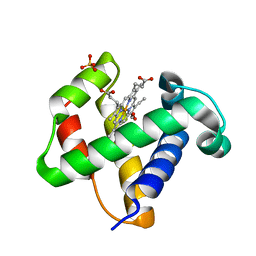 | | CRYSTAL STRUCTURE OF ThrE11Val mutant of THE NERVE TISSUE MINI-HEMOGLOBIN FROM THE NEMERTEAN WORM CEREBRATULUS LACTEUS | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Ascenzi, P, Geuens, E, Dewilde, S, Moens, L, Bolognesi, M, Riggs, A, Hale, A, Deng, P, Nienhaus, G.U, Olson, J.S, Nienhaus, K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thr-E11 Regulates O2 Affinity in Cerebratulus Lacteus Mini-Hemoglobin
J.Biol.Chem., 279, 2004
|
|
1V08
 
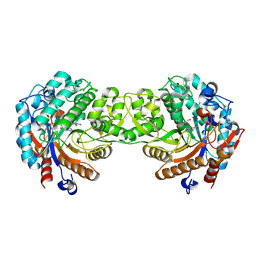 | | Crystal structure of the Zea maze beta-glucosidase-1 in complex with gluco-tetrazole | | Descriptor: | BETA-GLUCOSIDASE, NOJIRIMYCINE TETRAZOLE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V0A
 
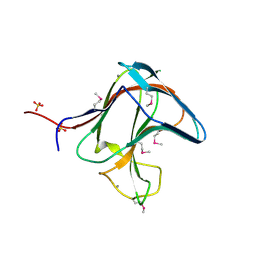 | | Family 11 Carbohydrate-Binding Module of cellulosomal cellulase Lic26A-Cel5E of Clostridium thermocellum | | Descriptor: | CALCIUM ION, ENDOGLUCANASE H, SULFATE ION | | Authors: | Carvalho, A.L, Romao, M.J, Goyal, A, Prates, J.A.M, Pires, V.M.R, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accomodates Beta-1,4- and Beta-1,3-1,4-Mixed Linked Glucans at a Single Binding Site
J.Biol.Chem., 279, 2004
|
|
1V0B
 
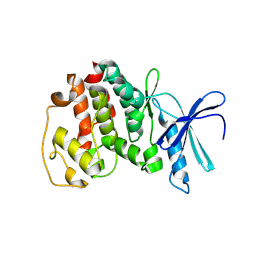 | | Crystal structure of the t198a mutant of pfpk5 | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-03-26 | | Release date: | 2004-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
1V0C
 
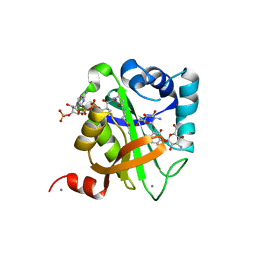 | | Structure of AAC(6')-Ib in complex with Kanamycin C and AcetylCoA. | | Descriptor: | AAC(6')-IB, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
1V0D
 
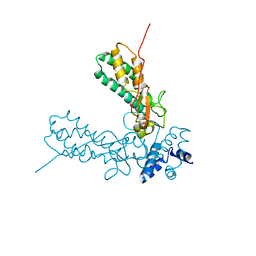 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
1V0E
 
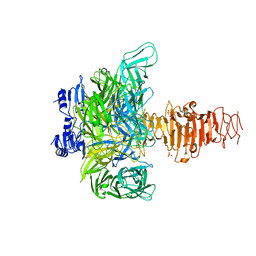 | | Endosialidase of Bacteriophage K1F | | Descriptor: | ENDO-ALPHA-SIALIDASE, PHOSPHATE ION | | Authors: | Stummeyer, K, Dickmanns, A, Muehlenhoff, M, Gerady-Schahn, R, Ficner, R. | | Deposit date: | 2004-03-28 | | Release date: | 2004-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Polysialic Acid-Degrading Endosialidase of Bacteriophage K1F
Nat.Struct.Mol.Biol., 12, 2005
|
|
1V0F
 
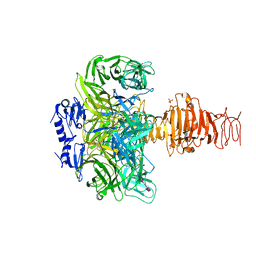 | | Endosialidase of Bacteriophage K1F in complex with oligomeric alpha-2,8-sialic acid | | Descriptor: | ENDO-ALPHA-SIALIDASE, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid, ... | | Authors: | Stummeyer, K, Dickmanns, A, Muehlenhoff, M, Gerady-Schahn, R, Ficner, R. | | Deposit date: | 2004-03-28 | | Release date: | 2004-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Polysialic Acid-Degrading Endosialidase of Bacteriophage K1F
Nat.Struct.Mol.Biol., 12, 2005
|
|
1V0H
 
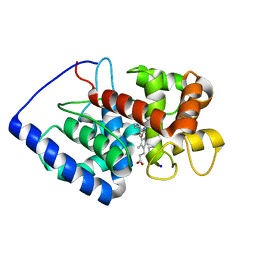 | | ASCOBATE PEROXIDASE FROM SOYBEAN CYTOSOL IN COMPLEX WITH SALICYLHYDROXAMIC ACID | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SALICYLHYDROXAMIC ACID, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Salicylhydroxamic Acid Complex
Biochemistry, 43, 2004
|
|
1V0J
 
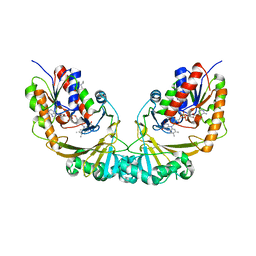 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
1V0K
 
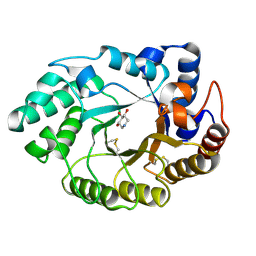 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki S, J, Withers, G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0L
 
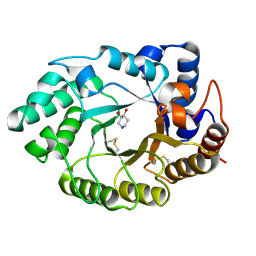 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0M
 
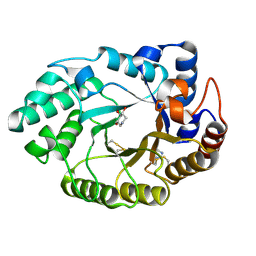 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
