6TNB
 
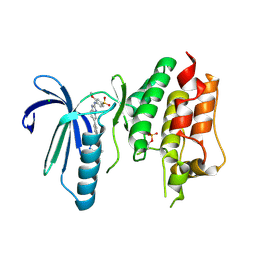 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 41 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, CHLORIDE ION, Dual specificity protein kinase TTK | | Authors: | Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Marquardt, T, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
5XG9
 
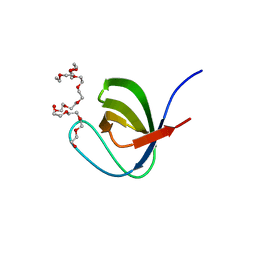 | | Crystal Structure of PEG-bound SH3 domain of Myosin IB from Entamoeba histolytica | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, PENTAETHYLENE GLYCOL, ... | | Authors: | Gautam, G, Gourinath, S. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the PEG-bound SH3 domain of myosin IB from Entamoeba histolytica reveals its mode of ligand recognition
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6TEW
 
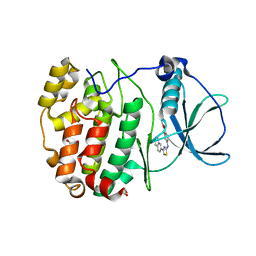 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor 27 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-naphthalen-2-yl-1,3-thiazol-2-yl)amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.082 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
5NQZ
 
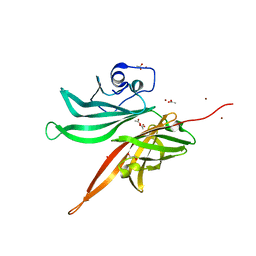 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
2PFO
 
 | | DNA Polymerase lambda in complex with DNA and dUPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA polymerase lambda, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
6RGG
 
 | | Photorhabdus laumondii lectin PLL2 in complex with O-methylated PGL-1-derived disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 3,6-O-dimethyl-D-glucose, 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
7UN8
 
 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
5NGT
 
 | | Crystal structure of human MTH1 in complex with inhibitor 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
7UNA
 
 | | SfSTING with cGAMP (masked) | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7CNZ
 
 | | Crystal structure of 10PE bound PSD from E. coli (2.70 A) | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, PHOSPHATE ION, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
5UOF
 
 | |
9HNC
 
 | | Crystal structure of potassium-independent L-asparaginase from Phaseolus vulgaris (PvAIII, PvAspG2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Loch, J.I, Pierog, I, Imiolczyk, B, Barciszewski, J, Marsolais, F, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Unique double-helical packing of protein molecules in the crystal of potassium-independent L-asparaginase from common bean.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7H1X
 
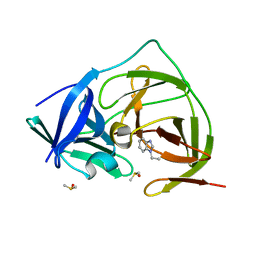 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z53833304 | | Descriptor: | 1-ethyl-1H-benzimidazole, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H1V
 
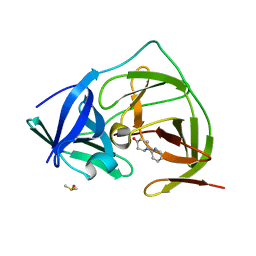 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z31113727 | | Descriptor: | 1-(pyridin-2-yl)piperidin-4-one, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H1O
 
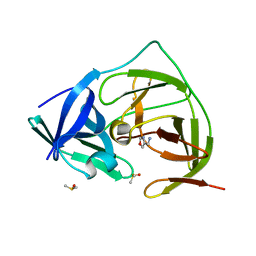 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1962142017 | | Descriptor: | 1,3-oxazol-4-ylmethanamine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
7H1P
 
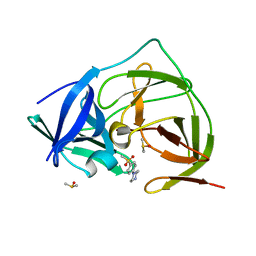 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z425338146 | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
6DP7
 
 | |
7H29
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1269184613 | | Descriptor: | 1,3-benzothiazole-6-sulfonamide, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
5ZA1
 
 | | Ligand complex of RORgt LBD | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-({[4-(ethylsulfonyl)phenyl]acetyl}amino)phenyl]-2-methyl-N-phenylpropanamide, DIMETHYLFORMAMIDE, ... | | Authors: | Yamamoto, S, Yamaguchi, H. | | Deposit date: | 2018-02-06 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of a potent orally bioavailable retinoic acid receptor-related orphan receptor-gamma-t (ROR gamma t) inhibitor, S18-000003.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5ZVX
 
 | |
5ZWA
 
 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium in complex with ADP, PL-linked to Lys233 via Schiff base in protomer A and the product (PLP) in protomer B | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
7BU3
 
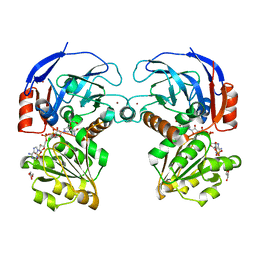 | | Structure of alcohol dehydrogenase YjgB in complex with NADP from Escherichia coli | | Descriptor: | ASPARTIC ACID, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nguyen, G.T, Kim, Y.-G, Ahn, J.-W, Chang, J.H. | | Deposit date: | 2020-04-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Molecules, 25, 2020
|
|
5HKG
 
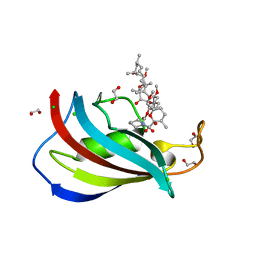 | | Total chemical synthesis, refolding and crystallographic structure of a fully active immunophilin: calstabin 2 (FKBP12.6). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Sirigu, S, Huet, T, Bacchi, M, Jullian, M, Fould, B, Ferry, G, Vuillard, L, Chavas, L, Puget, K, Nosjean, O, Boutin, J.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Total chemical synthesis, refolding, and crystallographic structure of fully active immunophilin calstabin 2 (FKBP12.6).
Protein Sci., 25, 2016
|
|
6RU2
 
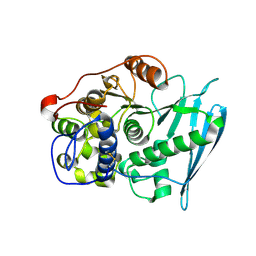 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
