1H7S
 
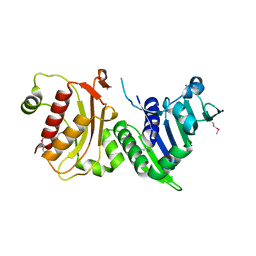 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1MST
 
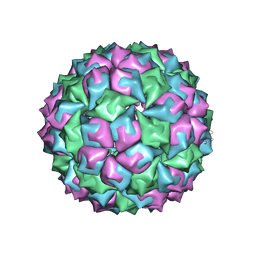 | |
1MSB
 
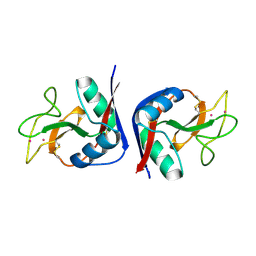 | |
1N1F
 
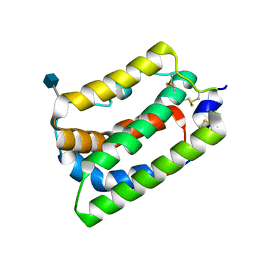 | | Crystal Structure of Human Interleukin-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, interleukin-19 | | Authors: | Chang, C, Magracheva, E, Kozlov, S, Fong, S, Tobin, G, Kotenko, S, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of interleukin-19 defines a new subfamily of helical cytokines
J.Biol.Chem., 278, 2003
|
|
1MTU
 
 | | FACTOR XA SPECIFIC INHIBITOR IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | (+)-2-[4-[(-1-ACETIMIDOYL-4-PIPERIDINYL)OXY]-3-(7-AMIDINO-2-NAPHTHYL)PROPIONIC ACID, CALCIUM ION, TRYPSIN | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of factor Xa specific inhibitors in complex with trypsin: structural grounds for inhibition of factor Xa and selectivity against thrombin.
FEBS Lett., 375, 1995
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MUD
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | Descriptor: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
1MWL
 
 | |
1MWP
 
 | | N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN | | Descriptor: | AMYLOID A4 PROTEIN | | Authors: | Rossjohn, J, Cappai, R, Feil, S.C, Henry, A, McKinstry, W.J, Galatis, D, Hesse, L, Multhaup, G, Beyreuther, K, Masters, C.L, Parker, M.W. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein.
Nat.Struct.Biol., 6, 1999
|
|
1MSW
 
 | |
1MWT
 
 | |
1MXF
 
 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1MZL
 
 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
1N2F
 
 | | CRYSTAL STRUCTURE OF P. AERUGINOSA OHR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Organic Hydroperoxide Resistance Protein | | Authors: | Lesniak, J, Barton, W.A, Nikolov, D.B. | | Deposit date: | 2002-10-22 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr
Embo J., 21, 2002
|
|
1MSK
 
 | | METHIONINE SYNTHASE (ACTIVATION DOMAIN) | | Descriptor: | ACETATE ION, COBALAMIN-DEPENDENT METHIONINE SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Dixon, M.M, Huang, S, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1996-08-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12.
Structure, 4, 1996
|
|
1MYM
 
 | |
1MVH
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1N2G
 
 | |
1GXS
 
 | | Crystal Structure of Hydroxynitrile Lyase from Sorghum bicolor in Complex with Inhibitor Benzoic Acid: a novel cyanogenic enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZOIC ACID, DECANOIC ACID, ... | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2002-04-11 | | Release date: | 2002-10-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Hydroxynitrile Lyase from Sorghum Bicolor in Complex with the Inhibitor Benzoic Acid: A Novel Cyanogenic Enzyme
Biochemistry, 41, 2002
|
|
1DEB
 
 | |
1MSA
 
 | |
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MSJ
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1MT0
 
 | | ATP-binding domain of hemolysin B from Escherichia coli | | Descriptor: | Hemolysin secretion ATP-binding protein, SULFATE ION | | Authors: | Schmitt, L, Benabdelhak, H, Blight, M.A, Holland, I.B, Stubbs, M.T. | | Deposit date: | 2002-09-20 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the nucleotide binding domain of the
ABC-transporter hemolysin B: identification of a variable region
within ABC helical domains
J.Mol.Biol., 330, 2003
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
