1Q63
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-Diamino-8-(1H-imidazol-2-ylsulfanylmethyl)-3H-quinazoline-4-one crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1PWZ
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with (R)-styrene oxide and chloride | | Descriptor: | CHLORIDE ION, R-STYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PXB
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1POA
 
 | |
1H6E
 
 | |
1PTH
 
 | | The Structural Basis of Aspirin Activity Inferred from the Crystal Structure of Inactivated Prostaglandin H2 Synthase | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Loll, P.J, Picot, D, Garavito, R.M. | | Deposit date: | 1995-04-11 | | Release date: | 1996-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase.
Nat.Struct.Biol., 2, 1995
|
|
1PUA
 
 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a Phosphorylated, 19-residue Histone H3 peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
1PUE
 
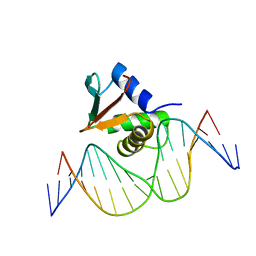 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
1Q21
 
 | |
1PVU
 
 | |
1PSI
 
 | |
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1Q67
 
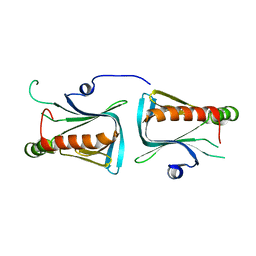 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1QHX
 
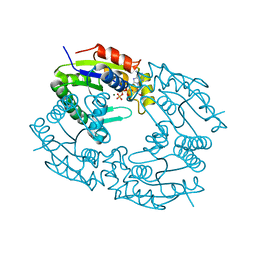 | |
1QIB
 
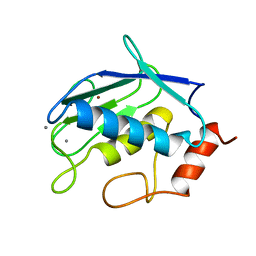 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1QIG
 
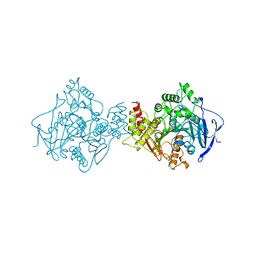 | |
1QIF
 
 | |
1QIR
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191Y MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QJQ
 
 | | FERRIC HYDROXAMATE RECEPTOR FROM ESCHERICHIA COLI (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, FERRIC HYDROXAMATE RECEPTOR, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-06-29 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
1QJC
 
 | |
1QKX
 
 | |
1QMR
 
 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | Descriptor: | MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
1QNB
 
 | | Crystal structure of the T(-25) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*TP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*AP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QL3
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the reduced state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QN4
 
 | | Crystal structure of the T(-24) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*TP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*AP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
