2CGU
 
 | | Identification of chemically diverse Chk1 inhibitors by receptor- based virtual screening | | Descriptor: | 2,2'-{[9-(HYDROXYIMINO)-9H-FLUORENE-2,7-DIYL]BIS(OXY)}DIACETIC ACID, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of Chemically Diverse Chk1 Inhibitors by Receptor-Based Virtual Screening.
Bioorg.Med.Chem., 14, 2006
|
|
2RJD
 
 | | Crystal structure of L3MBTL1 protein | | Descriptor: | Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2CJQ
 
 | |
2CI4
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I crystal form II | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CGX
 
 | | Identification of chemically diverse Chk1 inhibitors by receptor- based virtual screening | | Descriptor: | 2-[(6-AMINO-7H-PURIN-8-YL)THIO]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Chemically Diverse Chk1 Inhibitors by Receptor-Based Virtual Screening.
Bioorg.Med.Chem., 14, 2006
|
|
2CI3
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase crystal form I | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2BZT
 
 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
2BIA
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure G) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BZ6
 
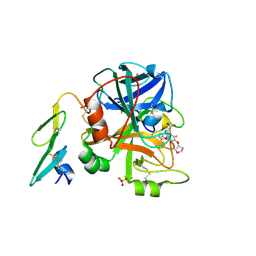 | | Orally available Factor7a inhibitor | | Descriptor: | (R)-(4-CARBAMIMIDOYL-PHENYLAMINO)-[5-ETHOXY-2-FLUORO-3-[(R)-TETRAHYDRO-FURAN-3-YLOXY]-PHENYL]-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Obst-Sander, U, Hilpert, K, Kuehne, H, Banner, D.W, Boehm, H.J, Stahl, M, Ackermann, J, Alig, L, Weber, L, Wessel, H.P, Riederer, M.A, Tschopp, T.B, Lave, T. | | Deposit date: | 2005-08-11 | | Release date: | 2006-02-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dose-Dependant Antithrombotic Activity of an Orally Active Tissue Factor/Factor Viia Inhibitor without Concomitant Enhancement of Bleeding Propensity.
Bioorg.Med.Chem., 14, 2006
|
|
2C1B
 
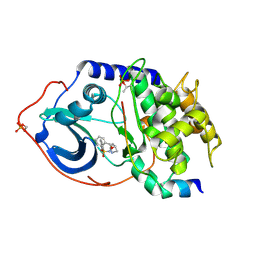 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2BCX
 
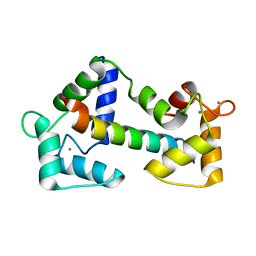 | | Crystal structure of calmodulin in complex with a ryanodine receptor peptide | | Descriptor: | CALCIUM ION, Calmodulin, Ryanodine receptor 1 | | Authors: | Maximciuc, A.A, Shamoo, Y, MacKenzie, K.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode.
Structure, 14, 2006
|
|
2BI9
 
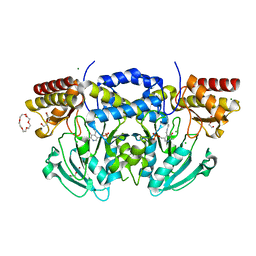 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure F) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BX6
 
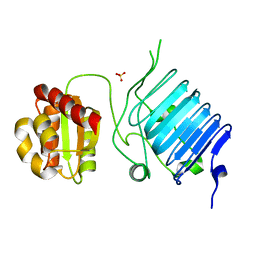 | | Crystal Structure of the human Retinitis Pigmentosa protein 2 (RP2) | | Descriptor: | SULFATE ION, XRP2 PROTEIN | | Authors: | Kuhnel, K, Veltel, S, Schlichting, I, Wittinghofer, A. | | Deposit date: | 2005-07-24 | | Release date: | 2006-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human Retinitis Pigmentosa 2 Protein and its Interaction with Arl3
Structure, 14, 2006
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
2RKK
 
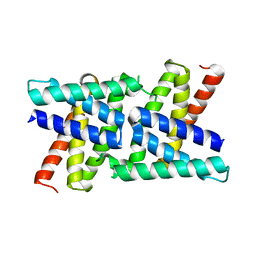 | |
5Z3Q
 
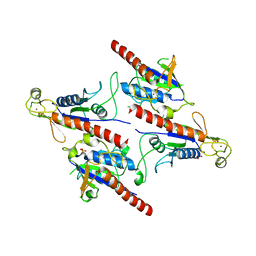 | | Crystal Structure of a Soluble Fragment of Poliovirus 2C ATPase (2.55 Angstrom) | | Descriptor: | PHOSPHATE ION, PV-2C, ZINC ION | | Authors: | Guan, H, Tian, J, Zhang, C, Qin, B, Cui, S. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Crystal structure of a soluble fragment of poliovirus 2CATPase
PLoS Pathog., 14, 2018
|
|
2D4Z
 
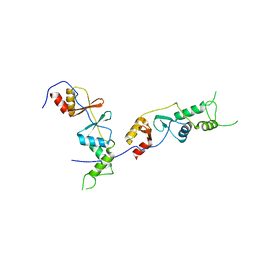 | |
5ZB1
 
 | | Monomeric crystal structure of orf57 from KSHV | | Descriptor: | ORF57, ZINC ION | | Authors: | Gao, Z.Q, Yuan, F, Lan, K, Dong, Y.H. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.
PLoS Pathog., 14, 2018
|
|
2D3M
 
 | | Pentaketide chromone synthase complexed with coenzyme A | | Descriptor: | COENZYME A, pentaketide chromone synthase | | Authors: | Morita, H, Kondo, S, Oguro, S, Noguchi, H, Sugio, S, Abe, I, Kohno, T. | | Deposit date: | 2005-09-29 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
2RJC
 
 | | Crystal structure of L3MBTL1 protein in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2D52
 
 | |
5ZQ1
 
 | | Crystal structure of spRlmCD with U1939loop RNA at 3.10 angstrom | | Descriptor: | RNA (5'-R(*AP*AP*AP*(MUM)P*UP*CP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Yu, H.L, Jiang, Y.Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
5YYF
 
 | |
5ZKB
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
2BF4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
