8UVL
 
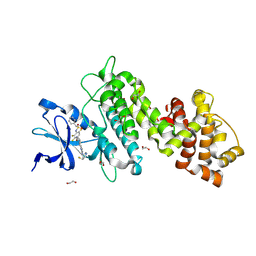 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
5JKN
 
 | | Crystal structure of deubiquitinase MINDY-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERCURY (II) ION, PHOSPHATE ION, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes.
Mol.Cell, 63, 2016
|
|
1EXR
 
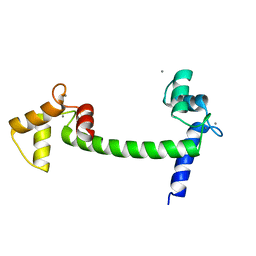 | |
9F6B
 
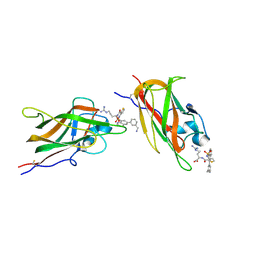 | | Human neuropilin-1 in a complex with a quinoline based antagonists | | Descriptor: | (2~{S})-2-[[3-[[6-[3-(aminomethyl)phenyl]quinolin-8-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | Authors: | Djordjevic, S, Selwood, D, Hubbard, P, Leonard, P, Mota, F. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Quinoline based neuropilin-1 antagonists exhibit a pure antagonist profile and block pain transmission
To Be Published
|
|
9B4Y
 
 | |
8R3R
 
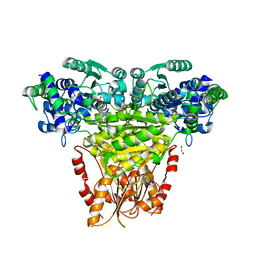 | | Transketolase from Streptococcus pneumoniae in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable transketolase, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biochemical, Bioinformatic, and Structural Comparisons of Transketolases and Position of Human Transketolase in the Enzyme Evolution.
Biochemistry, 63, 2024
|
|
8P54
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar MG-132. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8P5C
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6YUT
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with N-acyl functionalised cyclophellitol aziridine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
8JHQ
 
 | | Cryo-EM structure of human S1P transporter SPNS2 bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2,GlgA glycogen synthase | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8JHR
 
 | | Cryo-EM structure of human S1P transporter SPNS2 bound with an inhibitor 16d | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
8P2H
 
 | |
6J1M
 
 | |
6V3Q
 
 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | Descriptor: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
6BWL
 
 | | X-ray structure of Pal from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
6R7X
 
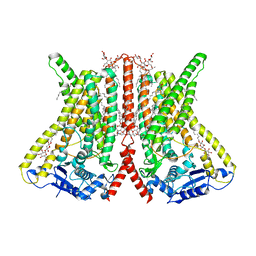 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
7CNZ
 
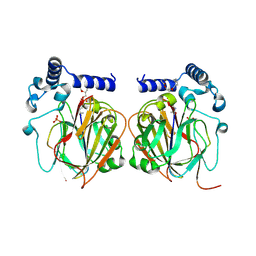 | | Crystal structure of 10PE bound PSD from E. coli (2.70 A) | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, PHOSPHATE ION, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6HOQ
 
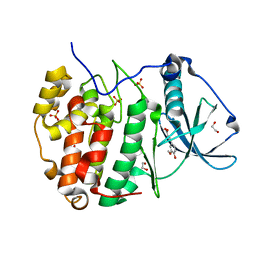 | | Human protein kinase CK2 alpha in complex with ferulic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Lolli, G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and cellular mechanism of protein kinase CK2 inhibition by deceptive curcumin.
Febs J., 287, 2020
|
|
6OJB
 
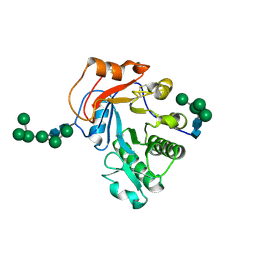 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
8TJI
 
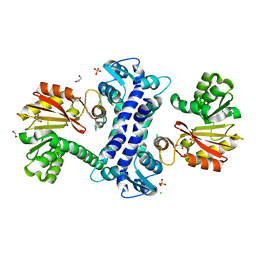 | | SAM-dependent methyltransferase RedM, apo | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
6R39
 
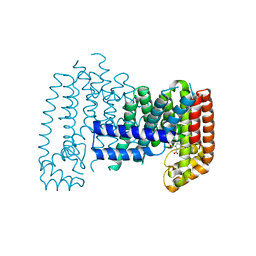 | |
6Z61
 
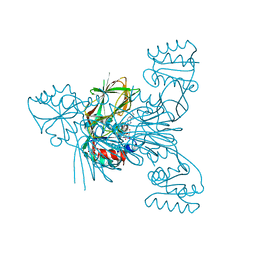 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[6-azanyl-8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-[6-(2-azanylethylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
