1BTS
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTU
 
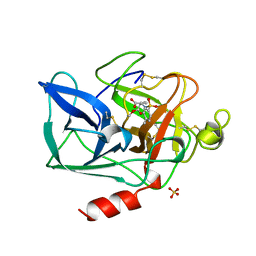 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4R)-1-TOLUENESULPHONYL-3-ETHYL-AZETIDIN-2-ONE-4-CARBOXYLIC ACID | | Descriptor: | (3R)-3-ethyl-N-[(4-methylphenyl)sulfonyl]-L-aspartic acid, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of elastase by N-sulfonylaryl beta-lactams: anatomy of a stable acyl-enzyme complex.
Biochemistry, 37, 1998
|
|
1BTV
 
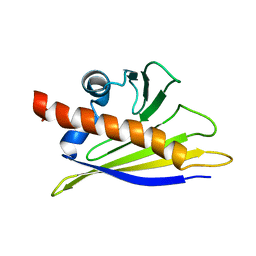 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1BTW
 
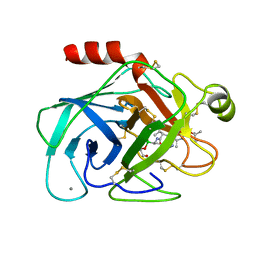 | |
1BTX
 
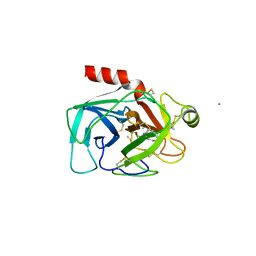 | |
1BTY
 
 | |
1BTZ
 
 | |
1BU1
 
 | | SRC FAMILY KINASE HCK SH3 DOMAIN | | Descriptor: | PROTEIN (HEMOPOIETIC CELL KINASE) | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1998-09-09 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef.
Biochemistry, 37, 1998
|
|
1BU2
 
 | |
1BU3
 
 | | REFINED CRYSTAL STRUCTURE OF CALCIUM-BOUND SILVER HAKE (PI 4.2) PARVALBUMIN AT 1.65 A. | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Richardson, R.C, Nelson, D.J, Royer, W.E, Harrington, D.J. | | Deposit date: | 1998-08-30 | | Release date: | 1999-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-Ray crystal structure and molecular dynamics simulations of silver hake parvalbumin (Isoform B).
Protein Sci., 9, 2000
|
|
1BU4
 
 | | RIBONUCLEASE 1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
1BU5
 
 | | X-RAY CRYSTAL STRUCTURE OF THE DESULFOVIBRIO VULGARIS (HILDENBOROUGH) APOFLAVODOXIN-RIBOFLAVIN COMPLEX | | Descriptor: | PROTEIN (FLAVODOXIN), RIBOFLAVIN, SULFATE ION | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Mccardle, P, Cunningham, P.D, Mayhew, S.G, Higgins, T.M. | | Deposit date: | 1998-09-12 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of the Desulfovibrio vulgaris (Hildenborough) apoflavodoxin-riboflavin complex.
Eur.J.Biochem., 258, 1998
|
|
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1BU7
 
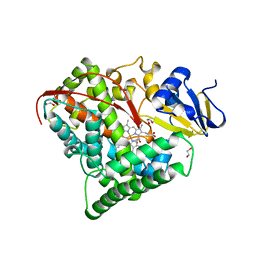 | | CRYOGENIC STRUCTURE OF CYTOCHROME P450BM-3 HEME DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYTOCHROME P450), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BU8
 
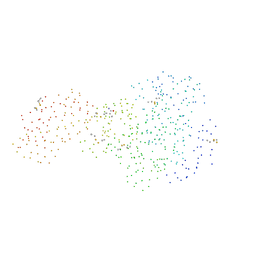 | | RAT PANCREATIC LIPASE RELATED PROTEIN 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PANCREATIC LIPASE RELATED PROTEIN 2) | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-09-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of rat pancreatic lipase-related protein 2.
J.Biol.Chem., 273, 1998
|
|
1BU9
 
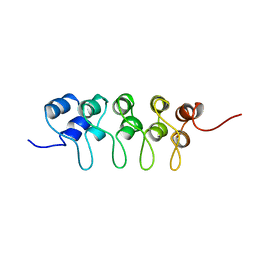 | | SOLUTION STRUCTURE OF P18-INK4C, 21 STRUCTURES | | Descriptor: | PROTEIN (CYCLIN-DEPENDENT KINASE 6 INHIBITOR) | | Authors: | Byeon, I.-J.L, Li, J, Tsai, M.-D. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A.
Biochemistry, 38, 1999
|
|
1BUA
 
 | |
1BUB
 
 | |
1BUC
 
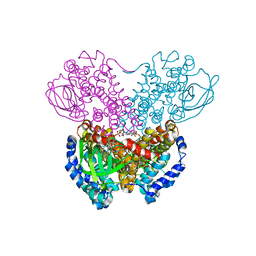 | | THREE-DIMENSIONAL STRUCTURE OF BUTYRYL-COA DEHYDROGENASE FROM MEGASPHAERA ELSDENII | | Descriptor: | ACETOACETYL-COENZYME A, BUTYRYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Djordjevic, S, Pace, C.P, Stankovich, M.T, Kim, J.J.P. | | Deposit date: | 1994-09-06 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii.
Biochemistry, 34, 1995
|
|
1BUD
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 5.0 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
1BUE
 
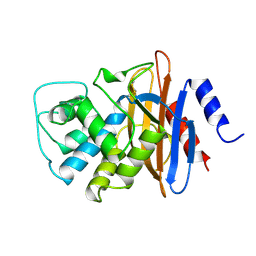 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
1BUF
 
 | |
1BUG
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES)-INHIBITOR COMPLEX WITH PHENYLTHIOUREA (PTU) | | Descriptor: | COPPER (II) ION, N-PHENYLTHIOUREA, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BUH
 
 | |
