8Q1C
 
 | | Substrate-free D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
8Q1A
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with inhibitor | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-pentyl-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q19
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 2-but-2-ynyl-1,1,3-tris(oxidanylidene)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q18
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-(2-phenylethyl)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q17
 
 | | Identification and optimisation of novel inhibitors of the Polyketide synthetase 13 thioesterase domain with antitubercular activity | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, GLYCEROL, Polyketide synthase Pks13, ... | | Authors: | Eadsforth, T.C, Punekar, A.S, Green, S.R, Baragana, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Identification and Optimization of Novel Inhibitors of the Polyketide Synthase 13 Thioesterase Domain with Antitubercular Activity.
J.Med.Chem., 66, 2023
|
|
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8Q0Q
 
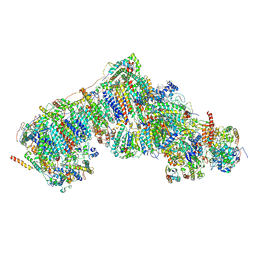 | | Outward-facing, slack proteoliposome complex I at 3.6 A. Initially purified in DDM | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q0P
 
 | |
8Q0O
 
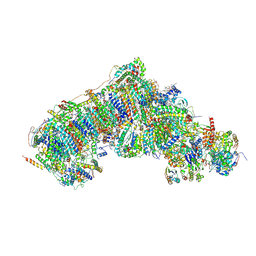 | | Outward-facing, open2 proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q0M
 
 | | Outward-facing, closed proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q0J
 
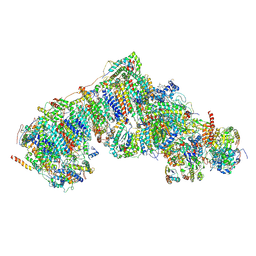 | | Inward-facing, slack proteoliposome complex I at 3.8 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q0F
 
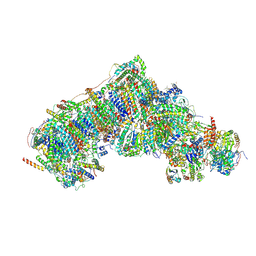 | | Inward-facing, open2 proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q0E
 
 | |
8Q0A
 
 | | Inward-facing, closed proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q05
 
 | |
8Q03
 
 | |
8PZK
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZ7
 
 | | crystal structure of VDR complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 57 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{R},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,8,9,9~{a}-hexahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-methyl-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
8PYY
 
 | |
8PYW
 
 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8PYA
 
 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure | | Descriptor: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Schofield, C.J. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure
To Be Published
|
|
8PY9
 
 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure | | Descriptor: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Schofield, C.J. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure
To Be Published
|
|
8PY8
 
 | | Isopenicillin N synthase in complex with Fe, ACdV and O2 after 2s O2 exposure | | Descriptor: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Schofield, C.J. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Isopenicillin N synthase in complex with Fe, ACdV and O2 after 2s O2 exposure
To Be Published
|
|
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8PXZ
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with natural substrate | | Descriptor: | (2~{R})-2-[[(2~{S},6~{R})-6,7-bis(azanyl)-2-[[(4~{R})-4,5-bis(azanyl)-5-oxidanylidene-pentanoyl]amino]-7-oxidanylidene-heptanoyl]amino]propanoic acid, (2~{R})-2-azanyl-~{N}'-[(2~{S},6~{R})-6,7-bis(azanyl)-1-oxidanyl-7-oxidanylidene-heptan-2-yl]pentanediamide, 1,2-ETHANEDIOL, ... | | Authors: | de Munnik, M, Schofield, C.J. | | Deposit date: | 2023-07-24 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with natural substrate
To Be Published
|
|
