8SC4
 
 | |
8SC1
 
 | |
9FNJ
 
 | | Half-closed CODH/ACS in the acetylated state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, CO-dehydrogenase, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-10 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
8SC2
 
 | |
8SC6
 
 | | Human OCT1 bound to thiamine in inward-open conformation | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Solute carrier family 22 member 1 | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of promiscuous substrate transport by Organic Cation Transporter 1.
Nat Commun, 14, 2023
|
|
9FRH
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with (R)-GABOB in the primed state | | Descriptor: | (R)-amino-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
7S3G
 
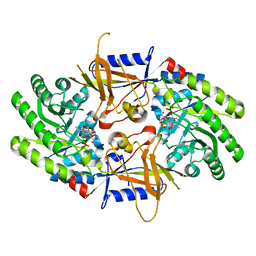 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
9FRF
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with (R)-GABOB in the desensitized state | | Descriptor: | (R)-amino-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
9FRG
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with (S)-GABOB in the desensitized state | | Descriptor: | (S)-amino-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
9FRB
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with CGP36742 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-azanylpropyl(butyl)phosphinic acid, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
9FRI
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with (S)-GABOB in the primed state | | Descriptor: | (S)-amino-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
9FWN
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00219 | | Descriptor: | 1-methyl-1-(phenylmethyl)urea, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00219
To Be Published
|
|
5GYH
 
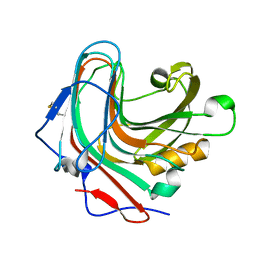 | |
5KTB
 
 | |
6VSR
 
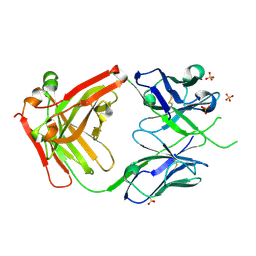 | |
7T9H
 
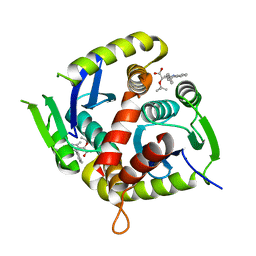 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
6VOR
 
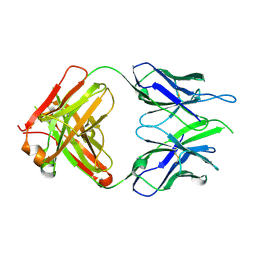 | | Crystal structure of macaque anti-HIV-1 antibody RM20E1 | | Descriptor: | GLYCINE, RM20E1 Fab heavy chain, RM20E1 Fab light chain | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
5AHO
 
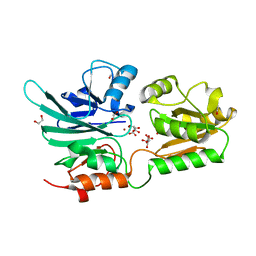 | | Crystal structure of human 5' exonuclease Apollo | | Descriptor: | 1,2-ETHANEDIOL, 5' EXONUCLEASE APOLLO, L(+)-TARTARIC ACID, ... | | Authors: | Allerston, C.K, Vollmar, M, Krojer, T, Pike, A.C.W, Newman, J.A, Carpenter, E, Quigley, A, Mahajan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
5LU7
 
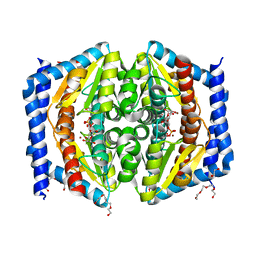 | | Heptose isomerase GmhA mutant - D61A | | Descriptor: | 1,2-ETHANEDIOL, 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, Phosphoheptose isomerase, ... | | Authors: | Vivoli, M, Harmer, N.J, Pang, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
6SSH
 
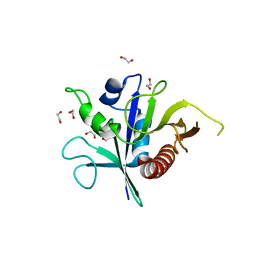 | | Structure of the TSC2 GAP domain | | Descriptor: | 1,2-ETHANEDIOL, GTPase activator-like protein | | Authors: | Hansmann, P, Kiontke, S, Kummel, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the TSC2 GAP Domain: Mechanistic Insight into Catalysis and Pathogenic Mutations.
Structure, 28, 2020
|
|
5BZW
 
 | |
2XSL
 
 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
1D41
 
 | |
9MZA
 
 | | Chemically Hijacked BCL6-TCIP3-p300 Complex | | Descriptor: | 1-{1-[5-({1-[5-chloro-4-({8-methoxy-1-methyl-3-[2-(methylamino)-2-oxoethoxy]-2-oxo-1,2-dihydroquinolin-6-yl}amino)pyrimidin-2-yl]piperidine-4-carbonyl}amino)pentanoyl]piperidin-4-yl}-3-[(6M)-7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, B-cell lymphoma 6 protein, Histone acetyltransferase p300 | | Authors: | Hinshaw, S.M, Gray, N.S, Nix, M.N, Gourisankar, S, Martinez, M, Crabtree, G.R. | | Deposit date: | 2025-01-22 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Bivalent Molecular Glue Linking Lysine Acetyltransferases to Oncogene-induced Cell Death.
Biorxiv, 2025
|
|
8RCV
 
 | | Crystal structure of HLA B*13:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen B alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
