5VRP
 
 | |
5VSK
 
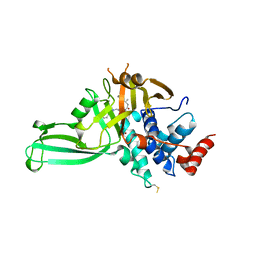 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
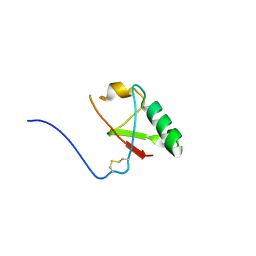
5VSX
 
 | |
5W2F
 
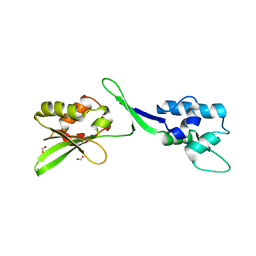 | | Crystal Structure of the C-terminal Domain of Human eIF2D at 1.4 A resolution | | Descriptor: | Eukaryotic translation initiation factor 2D, FORMIC ACID | | Authors: | Vaidya, A.T, Lomakin, I.B, Joseph, N.N, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
J. Mol. Biol., 429, 2017
|
|
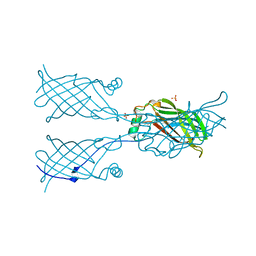
5W2K
 
 | |
5W2R
 
 | |
5VTL
 
 | |
3VT2
 
 | | Crystal structure of Ct1,3Gal43A in complex with isopropy-beta-D-thiogalactoside | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5W2X
 
 | |
5W31
 
 | |
3VT1
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactose | | Descriptor: | Ricin B lectin, beta-D-galactopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5W3B
 
 | |
1VEN
 
 | | Crystal Structure Analysis of Y164E/maltose of Bacilus cereus Beta-amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
5VVJ
 
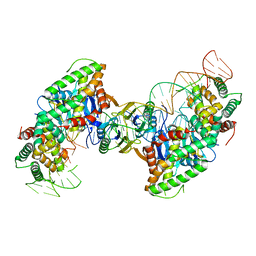 | | Cas1-Cas2 bound to half-site intermediate | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5W4R
 
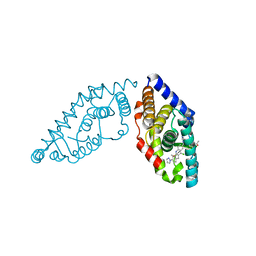 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | 1-{4-[(R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]piperidin-1-yl}ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W5B
 
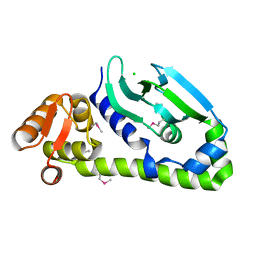 | | Crystal structure of Mycobacterium tuberculosis CRP-FNR family transcription factor Cmr (Rv1675c), truncated construct | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator Cmr | | Authors: | Cheung, J, Cassidy, M, Ginter, C, Ranganathan, S, Pata, D.J, McDonough, K.A. | | Deposit date: | 2017-06-14 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural features drive DNA binding properties of Cmr, a CRP family protein in TB complex mycobacteria.
Nucleic Acids Res., 46, 2018
|
|
5W5G
 
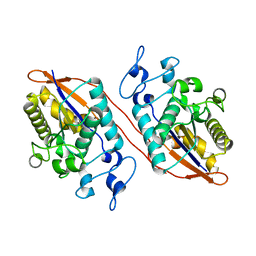 | | Structure of Human Sts-1 histidine phosphatase domain | | Descriptor: | Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
5VZO
 
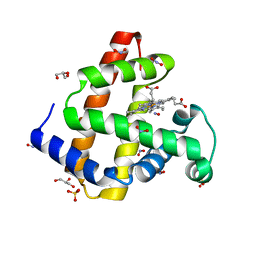 | | Sperm whale myoglobin H64A with nitric oxide | | Descriptor: | GLYCEROL, Myoglobin, NITRIC OXIDE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
5W6K
 
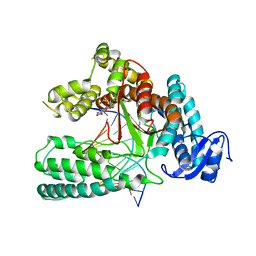 | | Structure of mutant Taq Polymerase incorporating unnatural base pairs Z:P | | Descriptor: | (1R)-1-[6-amino-5-(dihydroxyamino)-2-hydroxypyridin-3-yl]-1,4-anhydro-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-erythro-pentitol, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA (5'-D(P*(1WA)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Singh, I, Georgiadis, M.M. | | Deposit date: | 2017-06-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide.
Nucleic Acids Res., 46, 2018
|
|
5VZU
 
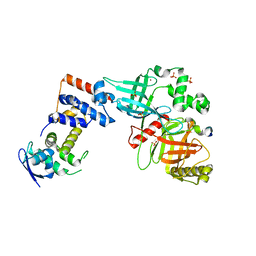 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W0G
 
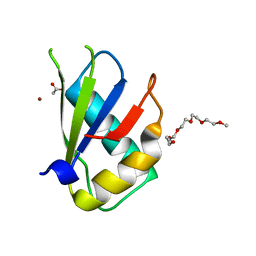 | | Structure of U2AF65 (U2AF2) RRM1 at 1.07 resolution | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, Splicing factor U2AF 65 kDa subunit, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2017-05-30 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs.
Biochemistry, 56, 2017
|
|
5W1N
 
 | |
1VGO
 
 | | Crystal Structure of Archaerhodopsin-2 | | Descriptor: | Archaerhodopsin 2, RETINAL, SULFATE ION, ... | | Authors: | Yoshimura, K, Enami, N, Murakami, M, Okumura, H, Ihara, K, Kouyama, T. | | Deposit date: | 2004-04-28 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps
J.Mol.Biol., 358, 2006
|
|
5W24
 
 | | Crystal Structure of 5C4 Fab | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5C4 Fab Heavy Chain, 5C4 Fab Light Chain | | Authors: | Battles, M.B, McLellan, J.S, Taher, N.M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
1VPD
 
 | | X-Ray Crystal Structure of Tartronate Semialdehyde Reductase [Salmonella Typhimurium LT2] | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, TARTRONATE SEMIALDEHYDE REDUCTASE | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
