1OWD
 
 | | Substituted 2-Naphthamidine inhibitors of urokinase | | Descriptor: | 6-[AMINO(IMINO)METHYL]-N-[(4R)-4-ETHYL-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL]-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OJZ
 
 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1OKB
 
 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OIL
 
 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1OSP
 
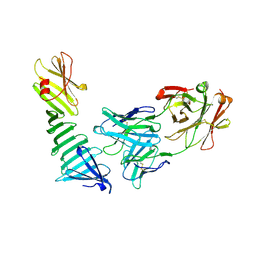 | |
1P0F
 
 | | Crystal Structure of the Binary Complex: NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) with the cofactor NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent ALCOHOL DEHYDROGENASE, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the vertebrate NADP(H)-dependent alcohol dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1P1G
 
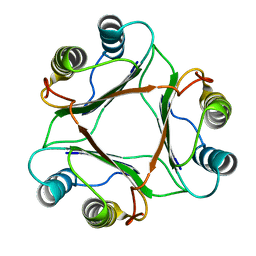 | |
1OWF
 
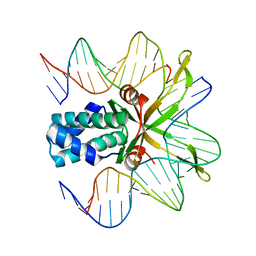 | | Crystal structure of a mutant IHF (BetaE44A) complexed with the native H' Site | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-28 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
1OYO
 
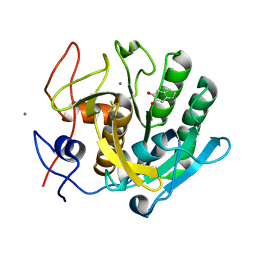 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
1P1U
 
 | |
1OJO
 
 | |
1P5Y
 
 | |
1P6K
 
 | | Rat neuronal NOS D597N mutant heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, SULFATE ION, yuaD protein | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-15 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
1OSA
 
 | |
1P92
 
 | | Crystal Structure of (H79A)DtxR | | Descriptor: | BETA-MERCAPTOETHANOL, Diphtheria toxin repressor | | Authors: | D'Aquino, J.A, Ringe, D. | | Deposit date: | 2003-05-08 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of the SRC homology domain 3-like fold.
J.Bacteriol., 185, 2003
|
|
1OM1
 
 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
1PAF
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
1OOY
 
 | | SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase, ... | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OPR
 
 | |
1OWG
 
 | | Crystal structure of WT IHF complexed with an altered H' site (T44A) | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*AP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-28 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
1OSU
 
 | |
1OY1
 
 | | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105 | | Descriptor: | PUTATIVE sigma cross-reacting protein 27A | | Authors: | Benach, J, Edstrom, W, Ma, L.C, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105
To be Published
|
|
1OSV
 
 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OYQ
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, SULFATE ION, Trypsin, ... | | Authors: | Nar, H. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition Promiscuity of Dual Specific Thrombin and Factor Xa Blood Coagulation Inhibitors
Structure, 9, 2001
|
|
