2VCH
 
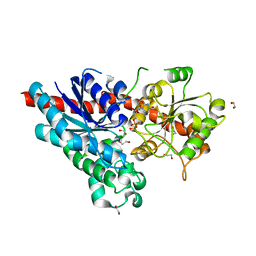 | | Characterization and engineering of the bifunctional N- and O- glucosyltransferase involved in xenobiotic metabolism in plants | | Descriptor: | 1,2-ETHANEDIOL, HYDROQUINONE GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Brazier-Hicks, M, Offen, W.A, Gershater, M.C, Revett, T.J, Lim, E.K, Bowles, D.J, Davies, G.J, Edwards, R. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization and Engineering of the Bifunctional N- and O-Glucosyltransferase Involved in Xenobiotic Metabolism in Plants.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1YAA
 
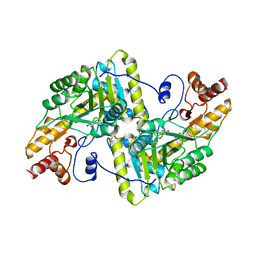 | |
2V1M
 
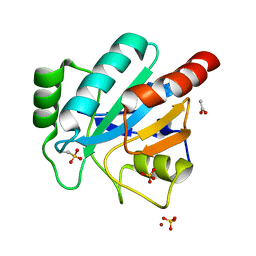 | | Crystal structure of Schistosoma mansoni glutathione peroxidase | | Descriptor: | ACETATE ION, GLUTATHIONE PEROXIDASE, LITHIUM ION, ... | | Authors: | Dimastrogiovanni, D, Miele, A.E, Angelucci, F, Boumis, G, Bellelli, A, Brunori, M. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Combining Crystallography and Molecular Dynamics: The Case of Schistosoma Mansoni Phospholipid Glutathione Peroxidase.
Proteins, 78, 2010
|
|
8V9Q
 
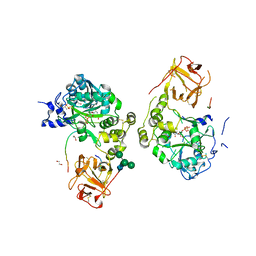 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
2VG9
 
 | | Crystal structure of Loop Swap mutant of Necallimastix patriciarum Xyn11A | | Descriptor: | ACETATE ION, BIFUNCTIONAL ENDO-1,4-BETA-XYLANASE A, CADMIUM ION | | Authors: | Vardakou, M, Dumon, C, Flint, J.E, Murray, J.W, Christakopoulos, P, Weiner, D.P, Juge, N, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Structural Basis for Substrate and Inhibitor Recognition in Eukaryotic Gh11 Xylanases.
J.Mol.Biol., 375, 2008
|
|
1YPT
 
 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YTS
 
 | | A LIGAND-INDUCED CONFORMATIONAL CHANGE IN THE YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | SULFATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Schubert, H.L, Stuckey, J.A, Fauman, E.B, Dixon, J.E, Saper, M.A. | | Deposit date: | 1995-04-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ligand-induced conformational change in the Yersinia protein tyrosine phosphatase.
Protein Sci., 4, 1995
|
|
7F0L
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES MONOMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein beta chain, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Nagashima, V.P, Kanno, R, Kawamura, S, Kikuchi, R, Ji, X.-C, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Nat Commun, 12, 2021
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1LW2
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
1MM9
 
 | | Streptavidin Mutant with Insertion of Fibronectin Hexapeptide, including RGD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, McDevitt, T.C, Nelson, K.E, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization and comparison of RGD cell-adhesion recognition sites engineered into streptavidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1T40
 
 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 at ph 5 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VNF
 
 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
1MMW
 
 | | Rat neuronal NOS heme domain with vinyl-L-NIO bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N5-(1-IMINO-3-BUTENYL)-L-ORNITHINE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
2VCE
 
 | | Characterization and engineering of the bifunctional N- and O- glucosyltransferase involved in xenobiotic metabolism in plants | | Descriptor: | 1,2-ETHANEDIOL, 2,4,5-trichlorophenol, HYDROQUINONE GLUCOSYLTRANSFERASE, ... | | Authors: | Brazier-Hicks, M, Offen, W.A, Gershater, M.C, Revett, T.J, Lim, E.K, Bowles, D.J, Davies, G.J, Edwards, R. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and Engineering of the Bifunctional N- and O-Glucosyltransferase Involved in Xenobiotic Metabolism in Plants.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1LWJ
 
 | | CRYSTAL STRUCTURE OF T. MARITIMA 4-ALPHA-GLUCANOTRANSFERASE/ACARBOSE COMPLEX | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, MODIFIED ACARBOSE PENTASACCHARIDE | | Authors: | Roujeinikova, A, Raasch, C, Sedelnikova, S, Liebl, W, Rice, D.W. | | Deposit date: | 2002-05-31 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA 4-ALPHA-GLUCANOTRANSFERASE AND ITS ACARBOSE COMPLEX:
IMPLICATIONS FOR SUBSTRATE SPECIFICITY AND CATALYSIS
J.Mol.Biol., 321, 2002
|
|
1Z1W
 
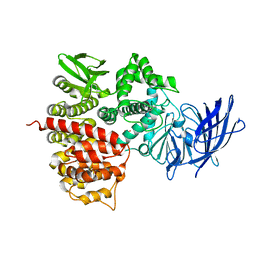 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1ZAI
 
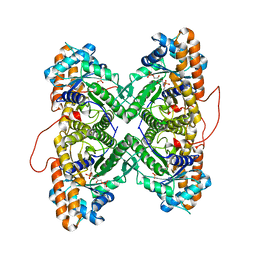 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from rabbit muscle | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1T7L
 
 | |
1K9Z
 
 | |
2V18
 
 | |
