5IQH
 
 | |
7EXW
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | Descriptor: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
5WB0
 
 | |
7EXU
 
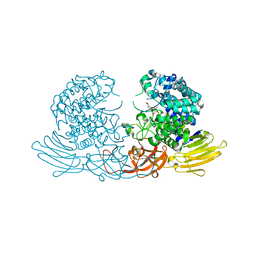 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | Descriptor: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5F2Y
 
 | |
5IHI
 
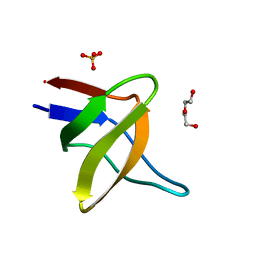 | |
5F3F
 
 | | Crystal structure of para-biphenyl-2-methyl-3'-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 3-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}-methyl-benzamide, Protein FimH | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2015-12-02 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Antivirulence C-Mannosides as Antibiotic-Sparing, Oral Therapeutics for Urinary Tract Infections.
J.Med.Chem., 59, 2016
|
|
5EUJ
 
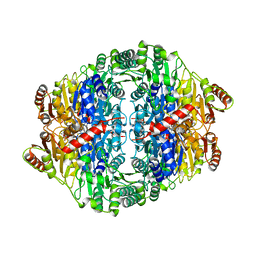 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5IPX
 
 | | Structure of ORF49 from KSHV | | Descriptor: | ORF49 protein, SULFATE ION | | Authors: | Hew, K, Nordlund, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Open Reading Frame 49 Protein Encoded by Kaposi's Sarcoma-Associated Herpesvirus.
J. Virol., 91, 2017
|
|
5IKG
 
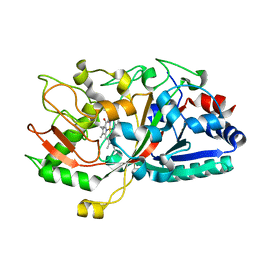 | |
5IQB
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GMPPNP, Magnesium, and Kanamycin A | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, KANAMYCIN A, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
5V80
 
 | |
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
5VAH
 
 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | Descriptor: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5F91
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FIX
 
 | |
5IT8
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
7F6Q
 
 | | Crystal structure of metal-citrate-binding mutant (S79A) protein (MctA) of ABC transporter in apo state | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETATE ION, ... | | Authors: | Kanaujia, S.P, Mandal, S.K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic insights into a novel Mg 2+ -citrate-binding protein from the ABC transporter superfamily.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5FB8
 
 | | Structure of Interleukin-16 bound to the 14.1 antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-IL-16 antibody 14.1 Fab domain Heavy Chain, ... | | Authors: | Hall, G, Cowan, R, Bayliss, R, Carr, M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a Potential Therapeutic Antibody Bound to Interleukin-16 (IL-16): MECHANISTIC INSIGHTS AND NEW THERAPEUTIC OPPORTUNITIES.
J.Biol.Chem., 291, 2016
|
|
5FBG
 
 | | S1 nuclease from Aspergillus oryzae, mutant D65N, in complex with phosphate, 2'-deoxycytidine and 2'-deoxyguanosine. | | Descriptor: | 2'-DEOXY-GUANOSINE, 2'-DEOXYCYTIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
7F6S
 
 | |
7F6T
 
 | |
