5JW9
 
 | |
3BLJ
 
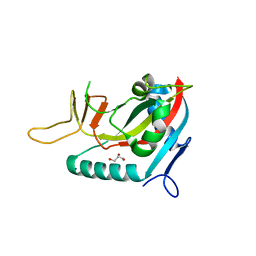 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ADP-ribose) polymerase 15, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
7XR2
 
 | |
7XR3
 
 | |
2G4C
 
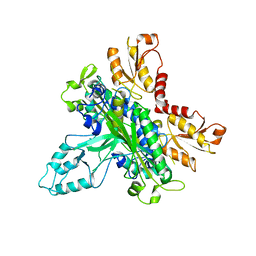 | | Crystal Structure of human DNA polymerase gamma accessory subunit | | Descriptor: | DNA polymerase gamma subunit 2 | | Authors: | Fan, L, Farr, C.L, Kaguni, L.S, Tainer, J.A. | | Deposit date: | 2006-02-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase
J.Mol.Biol., 358, 2006
|
|
7NXJ
 
 | | Crystal structure of human Cdk13/Cyclin K in complex with the inhibitor THZ531 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 13, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Anand, K, Greifenberg, A.K, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
7NXK
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with the inhibitor BSJ-01-175 | | Descriptor: | (E)-N-[4-[(1R,3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]cyclohexyl]oxyphenyl]-4-(dimethylamino)but-2-enamide, Cyclin-K, Cyclin-dependent kinase 12 | | Authors: | Anand, K, Dust, S, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
8SZU
 
 | |
2DB9
 
 | | Solution structure of the plus-3 domain of human KIAA0252 protein | | Descriptor: | Paf1/RNA polymerase II complex component | | Authors: | Yoneyama, M, Kigawa, T, Tochio, N, Nameki, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-06-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the plus-3 domain of human KIAA0252 protein
To be Published
|
|
6ALY
 
 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
2I53
 
 | | Crystal structure of Cyclin K | | Descriptor: | ACETATE ION, Cyclin K | | Authors: | Baek, K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2006-08-23 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Cyclin K, a Positive Regulator of Cyclin-dependent Kinase 9.
J.Mol.Biol., 366, 2007
|
|
6R0N
 
 | | Histone fold domain of AtNF-YB2/NF-YC3 in I2 | | Descriptor: | GLYCEROL, NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Bernardini, A, Fornara, F, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R0M
 
 | | Histone fold domain of AtNF-YB2/NF-YC3 in P212121 | | Descriptor: | NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
1QSS
 
 | | DDGTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-23 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8V7K
 
 | |
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
8V7F
 
 | | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T*())-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
2ZMI
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]pent-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZMH
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
8SZT
 
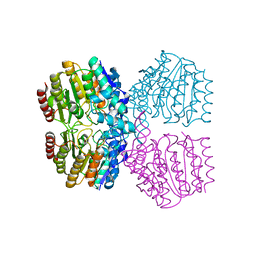 | | Structure of Kdac1 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, Histone deacetylase domain-containing protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of Kdac1, a Class II Deacetylase from the Multidrug-Resistant Pathogen Acinetobacter baumannii .
Biochemistry, 62, 2023
|
|
2XNF
 
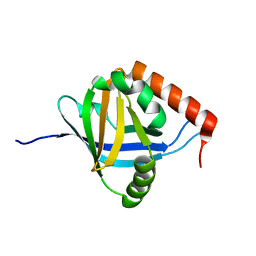 | | The Mediator Med25 activator interaction domain: Structure and cooperative binding of VP16 subdomains | | Descriptor: | MEDIATOR OF RNA POLYMERASE II TRANSCRIPTION SUBUNIT 25 | | Authors: | Vojnic, E, Mourao, A, Seizl, M, Simon, B, Wenzeck, L, Lariviere, L, Baumli, S, Meisterernst, M, Sattler, M, Cramer, P. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Vp16 Binding of the Mediator Med25 Activator Interaction Domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2ZMJ
 
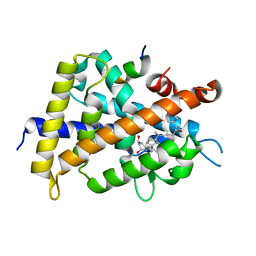 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
1QSY
 
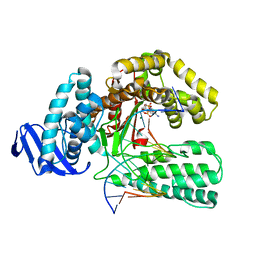 | | DDATP-Trapped closed ternary complex of the large fragment of DNA Polymerase I from thermus aquaticus | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, 5'-D(*AP*TP*TP*GP*CP*GP*CP*CP*TP*P*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2ZXM
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Ikura, T, Ito, N. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
