7D95
 
 | |
7D40
 
 | |
7D96
 
 | |
1NB9
 
 | | Crystal Structure of Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
7DW7
 
 | | Crystal Structure of N1051A mutant of Formylglycinamidine Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
8J28
 
 | | GK monomer complexes with G5P and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GK monomer complexes with G5P and ADP
To Be Published
|
|
8J27
 
 | | GK monomer complexes with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GK monomer complexes with ADP
To Be Published
|
|
3U0O
 
 | | The crystal structure of selenophosphate synthetase from E. coli | | Descriptor: | MAGNESIUM ION, Selenide, water dikinase | | Authors: | Noinaj, N, Wattanasak, R, Wally, J, Stadtman, T, Buchanan, S.K. | | Deposit date: | 2011-09-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the catalytic mechanism of Escherichia coli selenophosphate synthetase.
J.Bacteriol., 194, 2012
|
|
1T4A
 
 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
3A2K
 
 | | Crystal structure of TilS complexed with tRNA | | Descriptor: | bacterial tRNA, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
2D7J
 
 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
1TWJ
 
 | |
3Q7D
 
 | | Structure of (R)-naproxen bound to mCOX-2. | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Marnett, L.J. | | Deposit date: | 2011-01-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
4OIO
 
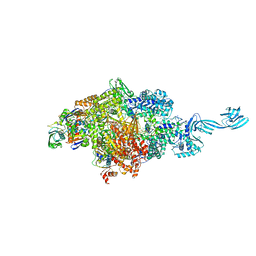 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4Q5S
 
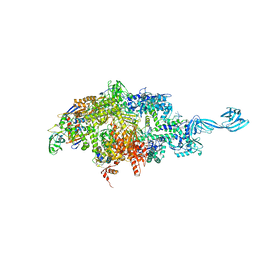 | | Thermus thermophilus RNA polymerase initially transcribing complex containing 6-mer RNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*GP*CP*AP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
1CT9
 
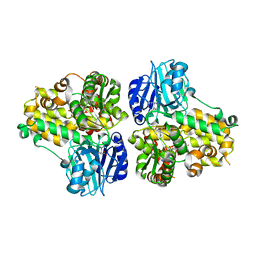 | | CRYSTAL STRUCTURE OF ASPARAGINE SYNTHETASE B FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE SYNTHETASE B, CHLORIDE ION, ... | | Authors: | Larsen, T.M, Boehlein, S.K, Schuster, S.M, Richards, N.G.J, Thoden, J.B, Holden, H.M, Rayment, I. | | Deposit date: | 1999-08-20 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product.
Biochemistry, 38, 1999
|
|
7LT2
 
 | |
5HM3
 
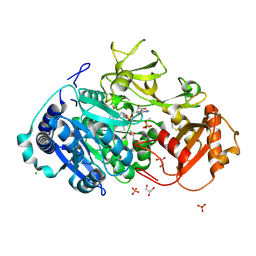 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
8B8T
 
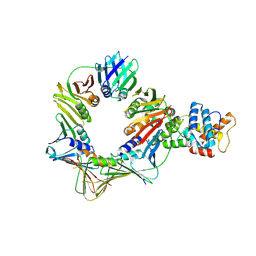 | | Open conformation of the complex of DNA ligase I on PCNA and DNA in the presence of ATP | | Descriptor: | DNA ligase 1, Proliferating cell nuclear antigen | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
7YC6
 
 | |
5ICR
 
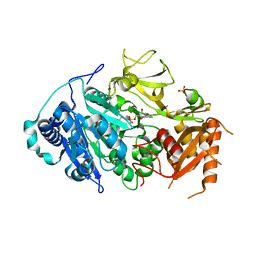 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-23 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
3OLT
 
 | |
3OLU
 
 | | X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of R513H murine COX-2 | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Malkowski, M.G. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structural basis of endocannabinoid oxygenation by cyclooxygenase-2.
J.Biol.Chem., 286, 2011
|
|
3ZFH
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | CHLORIDE ION, INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | Rao, V.A, Shepherd, S.M, Owen, R, Hunter, W.N. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Pseudomonas Aeruginosa Inosine 5'-Monophosphate Dehydrogenase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
8E9I
 
 | | Mycobacterial respiratory complex I, semi-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
