1NOJ
 
 | |
1NSQ
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Chiadmi, M, Morera, S, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
1NOK
 
 | |
1NOI
 
 | |
1NSP
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Chiadmi, M, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-18 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
1NZR
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1NPO
 
 | |
1OHD
 
 | | structure of cdc14 in complex with tungstate | | Descriptor: | CDC14B2 PHOSPHATASE, TUNGSTATE(VI)ION | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
1OGJ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 263 REPLACED BY PRO (L263P) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Martinez Julvez, M, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Involvement of the Pyrophosphate and the 2'-Phosphate Binding Regions of Ferredoxin-Nadp+ Reductase in Coenzyme Specificity
J.Biol.Chem., 278, 2003
|
|
1OGZ
 
 | | Crystal Structure Of 5-3-Ketosteroid Isomerase Mutants P39A Complexed With Equilenin From Pseudomonas Testosteroni | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Nam, G.H, Cha, S.-S, Yun, Y.S, Oh, Y.H, Hong, B.H, Lee, H.-S, Choi, K.Y. | | Deposit date: | 2003-05-20 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Conserved Cis-Pro39 Residue Plays a Crucial Role in the Proper Positioning of the Catalytic Base Asp38 in Ketosteroid Isomerase from Comamonas Testosteroni.
Biochem.J., 375, 2003
|
|
1OHT
 
 | | Peptidoglycan recognition protein LB | | Descriptor: | 1,2-ETHANEDIOL, CG14704 PROTEIN, L(+)-TARTARIC ACID, ... | | Authors: | Kim, M.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Peptidoglycan Recognition Protein Lb from Drosophila Melanogaster
Nat.Immunol., 4, 2003
|
|
1OFI
 
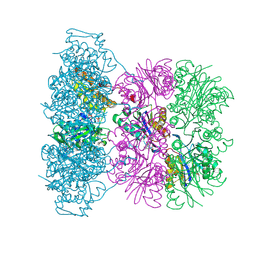 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFK
 
 | |
1OHS
 
 | |
1OGS
 
 | | human acid-beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Dvir, H, Harel, M, McCarthy, A.A, Toker, L, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structure of Human Acid-Beta-Glucosidase, the Defective Enzyme in Gaucher Disease
Embo Rep., 4, 2003
|
|
1OJN
 
 | |
1OK8
 
 | |
1OGF
 
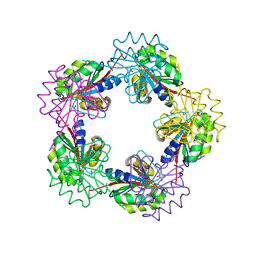 | | The Structure of Bacillus subtilis RbsD complexed with glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OLZ
 
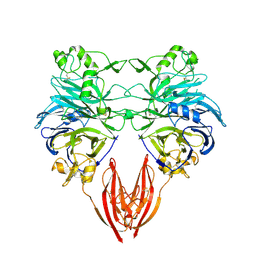 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1OHE
 
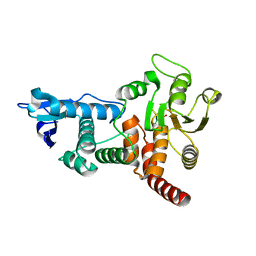 | | Structure of cdc14b phosphatase with a peptide ligand | | Descriptor: | CDC14B2 PHOSPHATASE, PEPTIDE LIGAND | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
1OHO
 
 | |
1OJP
 
 | |
1OON
 
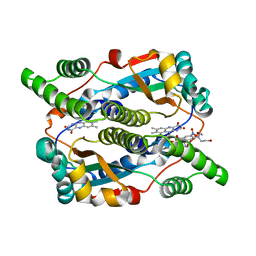 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN27217 | | Descriptor: | 2,4-DINITRO,5-[BIS(2-BROMOETHYL)AMINO]-N-(2',3'-DIOXOPROPYL)BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OKN
 
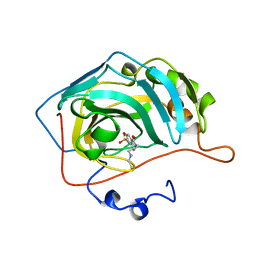 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1OFM
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B COMPLEXED TO CHONDROITIN 4-SULFATE TETRASACCHARIDE | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B, alpha-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-alpha-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Chondroitin B Lyase Complexed with Glycosaminoglycan Oligosaccharides Unravels a Calcium-Dependent Catalytic Machinery
J.Biol.Chem., 279, 2004
|
|
