3BEM
 
 | |
3BGU
 
 | |
4RGH
 
 | |
3BJA
 
 | |
4QRE
 
 | |
3BB9
 
 | |
3B7C
 
 | |
3W9P
 
 | |
3BDE
 
 | |
3WS3
 
 | | Crystal Structure of H-2D in complex with an insulin derived peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Kumar, P.R, Mukherjee, G, Samanta, D, DiLorenzo, T.P, Almo, S.C, Immune Function Network, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Compensatory mechanisms allow undersized anchor-deficient class I MHC ligands to mediate pathogenic autoreactive T cell responses
J. Immunol., 193, 2014
|
|
4QVU
 
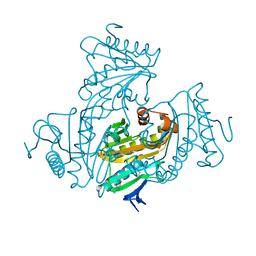 | |
4L8N
 
 | |
4R2I
 
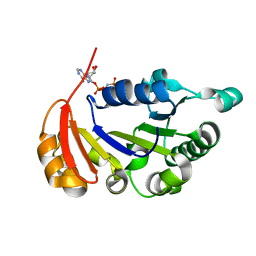 | | The Crystal Structure of STIV B204 complexed with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, STIV B204 ATPase, ZINC ION | | Authors: | Dellas, N, Nicolay, S.J, Young, M.J. | | Deposit date: | 2014-08-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Mutagenesis of Sulfolobus Turreted Icosahedral Virus B204 Reveals Essential Residues in the Virion-Associated DNA-Packaging ATPase.
J.Virol., 90, 2015
|
|
4L3R
 
 | |
4LER
 
 | |
3WNS
 
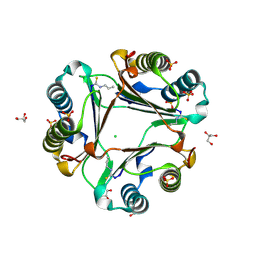 | | Allyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
4QRL
 
 | |
3BBJ
 
 | |
3BBY
 
 | |
4R5T
 
 | |
3B5O
 
 | |
3B81
 
 | |
3BFM
 
 | |
4LR4
 
 | |
4R24
 
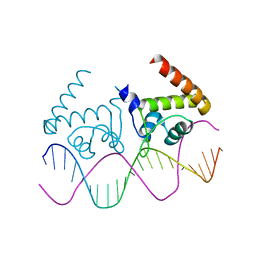 | | Complete dissection of B. subtilis nitrogen homeostatic circuitry | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), HTH-type transcriptional regulator TnrA | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
