6MM4
 
 | |
6D6X
 
 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
2OWI
 
 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
1LQC
 
 | | LAC REPRESSOR HEADPIECE (RESIDUES 1-56), NMR, 32 STRUCTURES | | Descriptor: | LAC REPRESSOR | | Authors: | Slijper, M, Bonvin, A.M.J.J, Boelens, R, Kaptein, R. | | Deposit date: | 1996-08-13 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator.
J.Mol.Biol., 259, 1996
|
|
2YTX
 
 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTV
 
 | | Solution structure of the fifth cold-shock domain of the human KIAA0885 protein (unr protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTY
 
 | | Solution structure of the fourth cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2OPV
 
 | |
7LSO
 
 | | L-Phenylseptin | | Descriptor: | L-Phenylseptin peptide | | Authors: | Munhoz, V.H, Verly, R.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of L-Phenylseptin in DPC-d38.
To Be Published
|
|
7LSP
 
 | |
3LEU
 
 | | HIGH RESOLUTION 1H NMR STUDY OF LEUCOCIN A IN DODECYLPHOSPHOCHOLINE MICELLES, 19 STRUCTURES (1:40 RATIO OF LEUCOCIN A:DPC) (0.1% TFA) | | Descriptor: | LEUCOCIN A | | Authors: | Gallagher, N.L.F, Sailer, M, Niemczura, W.P, Nakashima, T.T, Stiles, M.E, Vederas, J.C. | | Deposit date: | 1997-05-20 | | Release date: | 1997-11-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 36, 1997
|
|
6GMY
 
 | | Tc-DNA/RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*AP*AP*GP*CP*CP*GP*AP*G)-3'), Tc-DNA (5'-(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GPI
 
 | | Tc-DNA/DNA duplex | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*GP*CP*CP*GP*AP*G)-3'), tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-06-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
3GAT
 
 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
6RK3
 
 | | Solution structure of the ribosome Elongation Factor P (EF-P) from Staphylococcus aureus | | Descriptor: | Elongation factor P | | Authors: | Usachev, K, Fatkhullin, B, Gabdulkhakov, A, Khusainov, I, Golubev, A, Validov, S, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
1PFD
 
 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
6E98
 
 | |
6E9M
 
 | |
2RQ0
 
 | |
1SIS
 
 | | SPATIAL STRUCTURE OF INSECTOTOXIN I5A BUTHUS EUPEUS BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY (RUSSIAN) | | Descriptor: | SCORPION INSECTOTOXIN I5A | | Authors: | Arseniev, A.S, Kondakov, V.I, Lomize, A.L, Maiorov, V.N, Bystrov, V.F. | | Deposit date: | 1993-11-11 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the spatial structure of insectotoxin 15A from Buthus erpeus by (1)H-NMR spectroscopy data
Bioorg.Khim., 17, 1991
|
|
1K2H
 
 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
1H2O
 
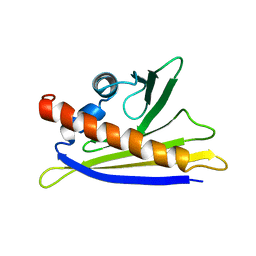 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
2RMQ
 
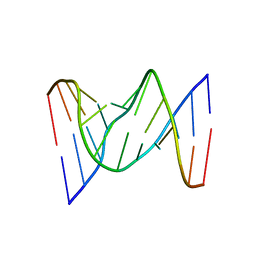 | | Solution structure of fully modified 4'-thioDNA with the sequence of d(CGCGAATTCGCG) | | Descriptor: | DNA (5'-D(*(C4S)P*(S4G)P*(C4S)P*(S4G)P*(S4A)P*(S4A)P*(T49)P*(T49)P*(C4S)P*(S4G)P*(C4S)P*(S4G))-3') | | Authors: | Matsugami, A, Ohyama, T, Inada, M, Katahira, M. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Unexpected A-form formation of 4'-thioDNA in solution, revealed by NMR, and the implications as to the mechanism of nuclease resistance
Nucleic Acids Res., 36, 2008
|
|
