1JP0
 
 | |
1JQ9
 
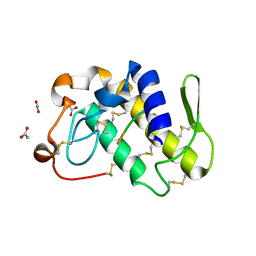 | | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2 | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution
J.BIOL.CHEM., 277, 2002
|
|
1JQI
 
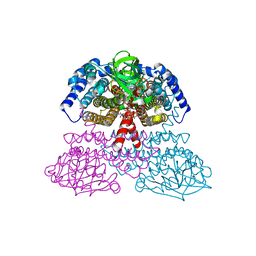 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
1JOA
 
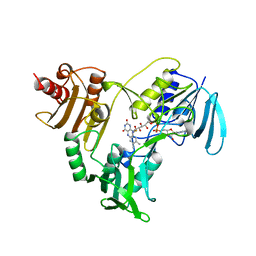 | | NADH PEROXIDASE WITH CYSTEINE-SULFENIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE | | Authors: | Yeh, J.I, Claiborne, A.C, Hol, W.G.J. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 A resolution.
Biochemistry, 35, 1996
|
|
1YSL
 
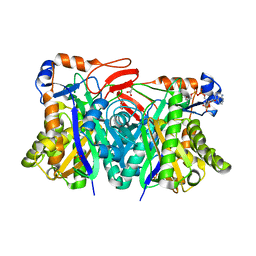 | | Crystal structure of HMG-CoA synthase from Enterococcus faecalis with AcetoAcetyl-CoA ligand. | | Descriptor: | ACETOACETIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Steussy, C.N, Vartia, A.A, Burgner II, J.W, Sutherlin, A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2005-02-08 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structures of HMG-CoA Synthase from Enterococcus faecalis and a Complex with Its Second Substrate/Inhibitor Acetoacetyl-CoA.
Biochemistry, 44, 2005
|
|
1YMU
 
 | |
1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1JS3
 
 | | Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa | | Descriptor: | CARBIDOPA, DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
1YBV
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
1YMV
 
 | |
1JU2
 
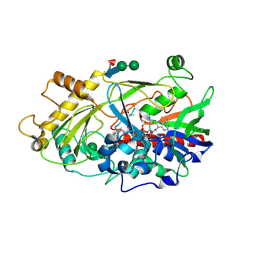 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
1YCP
 
 | |
1YTJ
 
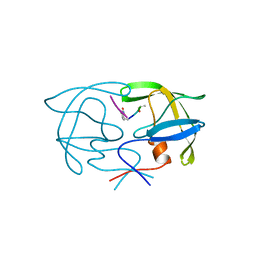 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | PEPTIDE PRODUCT, SIV PROTEASE | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YJC
 
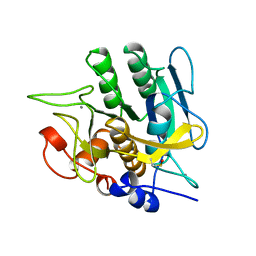 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 50% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YJA
 
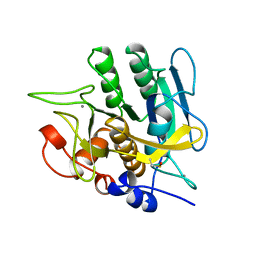 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 20% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YPN
 
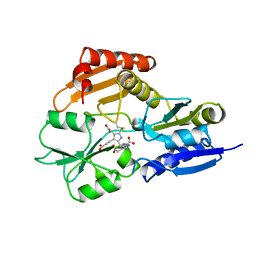 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 1998-06-26 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
1YTH
 
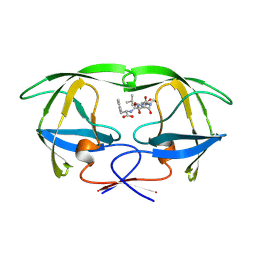 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1JVT
 
 | |
1YFO
 
 | |
1YAA
 
 | |
1YER
 
 | |
1YDV
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Velankar, S.S, Murthy, M.R.N. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design.
Structure, 5, 1997
|
|
1YPR
 
 | |
1YST
 
 | | STRUCTURE OF THE PHOTOCHEMICAL REACTION CENTER OF A SPHEROIDENE CONTAINING PURPLE BACTERIUM, RHODOBACTER SPHAEROIDES Y, AT 3 ANGSTROMS RESOLUTION | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, MANGANESE (II) ION, ... | | Authors: | Arnoux, B, Gaucher, J.F, Ducruix, A, Reiss-Husson, F. | | Deposit date: | 1994-12-07 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the photochemical reaction centre of a spheroidene-containing purple-bacterium, Rhodobacter sphaeroides Y, at 3 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1K1O
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, {[(1R)-2-((2S)-2-{[(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)AMINO]CARBONYL}PIPERIDINYL)-1-(CYCLOHEXYLMETHYL)-2-OXOETHYL]AMINO}ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
