7SHW
 
 | |
6VP9
 
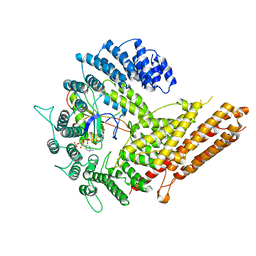 | | Cryo-EM structure of human NatB complex | | Descriptor: | CARBOXYMETHYL COENZYME *A, MDVFM peptide, N-alpha-acetyltransferase 20, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for N-terminal alpha-synuclein acetylation by human NatB.
Elife, 9, 2020
|
|
7SCI
 
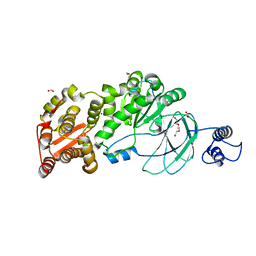 | | AM0627 metallopeptidase from Akkermansia muciniphila | | Descriptor: | CHLORIDE ION, FORMIC ACID, Peptidase M60 domain-containing protein, ... | | Authors: | Fernandez, D, Bertozzi, C.R, Shon, D.J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis of a mucin-selective metalloprotease from Akkermansia muciniphila alters substrate preferences.
J.Biol.Chem., 298, 2022
|
|
6VMD
 
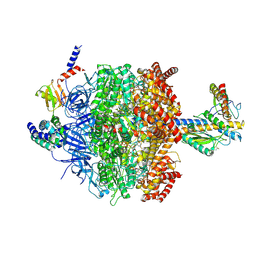 | | Chloroplast ATP synthase (C1, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VMG
 
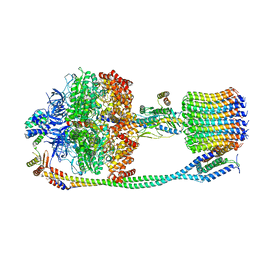 | | Chloroplast ATP synthase (O3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.46 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
3BRK
 
 | |
3C2I
 
 | | The Crystal Structure of Methyl-CpG Binding Domain of Human MeCP2 in Complex with a Methylated DNA Sequence from BDNF | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DGP*DAP*DAP*DGP*DAP*DAP*DTP*DTP*DCP*(5CM)P*DGP*DTP*DTP*DCP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DCP*DTP*DGP*DGP*DAP*DAP*(5CM)P*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DTP*DA)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, McNae, I.W, Schmiedeberg, L, Klose, R.J, Bird, A.P, Walkinshaw, M.D. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MeCP2 binding to DNA depends upon hydration at methyl-CpG
Mol.Cell, 29, 2008
|
|
8Q68
 
 | | Crystal structure of TEAD1-YBD in complex with irreversible compound SWTX-143 | | Descriptor: | Transcriptional enhancer factor TEF-1, ~{N}-[(3~{S})-5-azanyl-1-[4-(trifluoromethyl)phenyl]-3,4-dihydro-2~{H}-quinolin-3-yl]propanamide | | Authors: | Ciesielski, F, Spieser, S.A.H, Marchand, A, Gwaltney, S.L. | | Deposit date: | 2023-08-11 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Novel Irreversible TEAD Inhibitor, SWTX-143, Blocks Hippo Pathway Transcriptional Output and Causes Tumor Regression in Preclinical Mesothelioma Models.
Mol.Cancer Ther., 23, 2024
|
|
3C6A
 
 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
3BY9
 
 | |
7S2V
 
 | |
3C9B
 
 | | Crystal structure of SeMet Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Keck, J.L, Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
7SET
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1000 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7S2A
 
 | |
7S29
 
 | |
7S2F
 
 | |
7S2C
 
 | |
7S2G
 
 | |
7S30
 
 | |
7S33
 
 | |
7S28
 
 | |
7S2D
 
 | |
7S2B
 
 | |
7S32
 
 | |
7S34
 
 | |
