2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
1Z2M
 
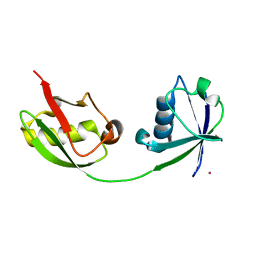 | | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | | Descriptor: | OSMIUM 4+ ION, interferon, alpha-inducible protein (clone IFI-15K) | | Authors: | Narasimhan, J, Wang, M, Fu, Z, Klein, J.M, Haas, A.L, Kim, J.J. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Interferon-induced Ubiquitin-like Protein ISG15.
J.Biol.Chem., 280, 2005
|
|
3REN
 
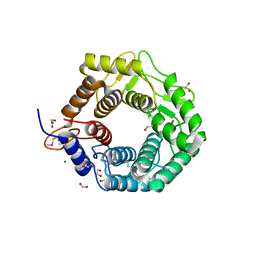 | | CPF_2247, a novel alpha-amylase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase, family 8, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Boraston, A.B. | | Deposit date: | 2011-04-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Proteins, 79, 2011
|
|
1KHO
 
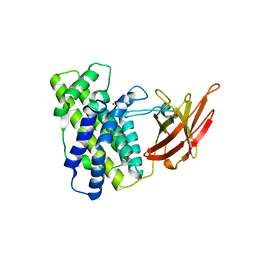 | | Crystal Structure Analysis of Clostridium perfringens alpha-Toxin Isolated from Avian Strain SWCP | | Descriptor: | ZINC ION, alpha-toxin | | Authors: | Justin, N, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The first strain of Clostridium perfringens isolated from an avian source has an alpha-toxin with divergent structural and kinetic properties.
Biochemistry, 41, 2002
|
|
2YHJ
 
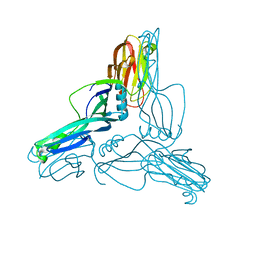 | | Clostridium perfringens Enterotoxin at 4.0 Angstrom Resolution | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, McClane, B.A, Basak, A.K. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
1M13
 
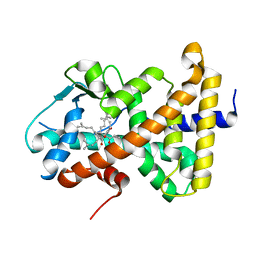 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
3KK7
 
 | |
5XCC
 
 | |
5XCB
 
 | |
5YFK
 
 | |
1GYG
 
 | |
5WQW
 
 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | Authors: | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | Deposit date: | 2016-11-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
2O61
 
 | |
4D70
 
 | | Structural, biophysical and biochemical analyses of a Clostridium perfringens Sortase D5 transpeptidase | | Descriptor: | SORTASE FAMILY PROTEIN | | Authors: | Suryadinata, R, Seabrook, S, Adams, T.E, Nuttall, S.D, Peat, T.S. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Biochemical Analyses of a Clostridium Perfringens Sortase D Transpeptidase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LPL
 
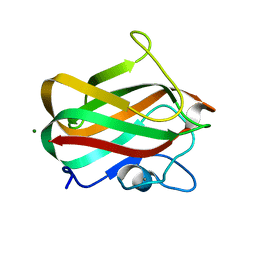 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
1UYJ
 
 | | Clostridium perfringens epsilon toxin shows structural similarity with the pore forming toxin aerolysin | | Descriptor: | EPSILON-TOXIN, URANIUM ATOM | | Authors: | Cole, A.R, Gibert, M, Poppoff, M, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clostridium Perfringens Epsilon-Toxin Shows Structural Similarity to the Pore-Forming Toxin Aerolysin
Nat.Struct.Mol.Biol., 11, 2004
|
|
4LKS
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Kirlin, A.C, Furness, H.S, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
2O4E
 
 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
5B23
 
 | |
4H56
 
 | | Crystal structure of the Clostridium perfringens NetB toxin in the membrane inserted form | | Descriptor: | Necrotic enteritis toxin B | | Authors: | Savva, C.G, Fernandes da Costa, S.P, Bokori-Brown, M, Naylor, C, Cole, A.R, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture and Functional Analysis of NetB, a Pore-forming Toxin from Clostridium perfringens.
J.Biol.Chem., 288, 2013
|
|
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2BF6
 
 | |
