6X3I
 
 | | NNAS Fc mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose | | Authors: | Wei, R, Zhou, Y.F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Engineered Fc-glycosylation switch to eliminate antibody effector function.
Mabs, 12, 2020
|
|
4WKH
 
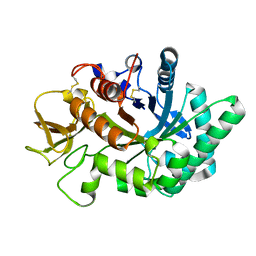 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WU0
 
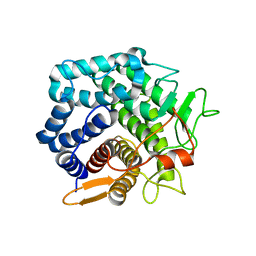 | | Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 | | Descriptor: | Similar to yteR (Bacilus subtilis) | | Authors: | Germane, K.L, Servinsky, M.D, Gerlach, E.S, Sund, C.J, Hurley, M.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Clostridium acetobutylicum ATCC 824 glycoside hydrolase from CAZy family GH105.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6CXD
 
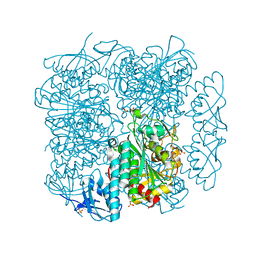 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | Descriptor: | Peptidase B, SULFATE ION | | Authors: | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6XTK
 
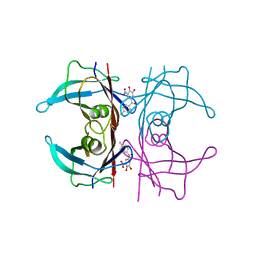 | | Y114C Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
6HIL
 
 | |
6HIK
 
 | |
6DXQ
 
 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
6RA6
 
 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
8CF4
 
 | |
8CHH
 
 | |
8CHG
 
 | |
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
8DFZ
 
 | |
1KTP
 
 | | Crystal structure of c-phycocyanin of synechococcus vulcanus at 1.6 angstroms | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, ... | | Authors: | Adir, N, Dobrovetsky, E, Lerner, N. | | Deposit date: | 2002-01-17 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structure of c-phycocyanin from the cyanobacterium Synechococcus vulcanus at 1.6 A: insights into the role of solvent molecules in thermal stability and co-factor structure
Biochim.Biophys.Acta, 1556, 2002
|
|
5JTW
 
 | | Crystal structure of complement C4b re-refined using iMDFF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4-A | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6CQA
 
 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6PBR
 
 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | Authors: | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6T9A
 
 | | Crystal structrue of RSL W31FW76F lectin mutant in complex with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, Fucose-binding lectin protein, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
6CW7
 
 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CYV
 
 | | E. coli DHFR ternary complex with NADP and dihydrofolate | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6T99
 
 | |
6T9B
 
 | | Crystal structrue of RSL W31A lectin mutant in complex with alpha-methylfucoside | | Descriptor: | Fucose-binding lectin protein, GLYCINE, methyl alpha-L-fucopyranoside | | Authors: | Houser, J, Komarek, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
6CXK
 
 | | E. coli DHFR substrate complex with Dihydrofolate | | Descriptor: | CHLORIDE ION, DIHYDROFOLIC ACID, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420314 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(S(-)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-31 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
