5VWE
 
 | |
6OBI
 
 | | Remarkable rigidity of the single alpha-helical domain of myosin-VI revealed by NMR spectroscopy | | Descriptor: | Myosin-VI | | Authors: | Barnes, A, Shen, Y, Ying, J, Takagi, Y, Torchia, D.A, Sellers, J, Bax, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Remarkable Rigidity of the Single alpha-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
9CJA
 
 | |
6QYS
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B | | Descriptor: | DBB-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6G81
 
 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | Descriptor: | Hydrogenase maturation factor HypA, ZINC ION | | Authors: | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | Descriptor: | CEMP1-p1 | | Authors: | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
1B2I
 
 | |
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
1B4C
 
 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | Descriptor: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | Authors: | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
8B4R
 
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, monomeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
6HD2
 
 | |
5JPX
 
 | |
2QMT
 
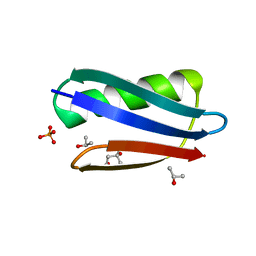 | | Crystal Polymorphism of Protein GB1 Examined by Solid-state NMR and X-ray Diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOPROPYL ALCOHOL, Immunoglobulin G-binding protein G, ... | | Authors: | Frericks Schmidt, H.L, Sperling, L.J, Gao, Y.G, Wylie, B.J, Boettcher, J.M, Wilson, S.R, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Polymorphism of Protein GB1 Examined by Solid-State NMR Spectroscopy and X-ray Diffraction.
J.Phys.Chem.B, 111, 2007
|
|
6MM4
 
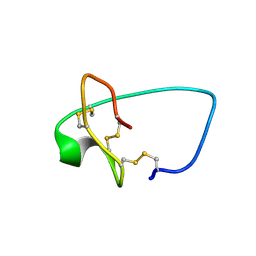 | |
5WT7
 
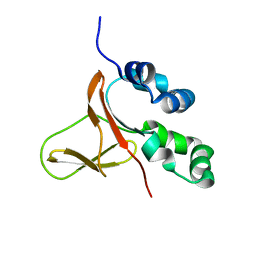 | | FAS1-IV domain of Human Periostin | | Descriptor: | Periostin | | Authors: | Yun, H, Lee, C.W. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C, and 15N resonance assignments of FAS1-IV domain of human periostin, a component of extracellular matrix proteins.
Biomol NMR Assign, 12, 2018
|
|
5XBO
 
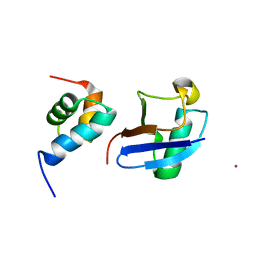 | | Lanthanoid tagging via an unnatural amino acid for protein structure characterization | | Descriptor: | Polyubiquitin-B, TERBIUM(III) ION, UV excision repair protein RAD23 homolog A | | Authors: | Jiang, W, Gu, X, Dong, X, Tang, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Lanthanoid tagging via an unnatural amino acid for protein structure characterization
J. Biomol. NMR, 67, 2017
|
|
6QM1
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B (Lan8,11) analogue | | Descriptor: | DAL-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QTF
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6S0N
 
 | | A9 peptide derived from Herceptin fab binding region | | Descriptor: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | Authors: | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
6ZFV
 
 | |
6E4J
 
 | | Solution NMR Structure of Protein PF2048.1 | | Descriptor: | Uncharacterized protein PF2048.1 | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
6J12
 
 | |
1H9O
 
 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AUD
 
 | | U1A-UTRRNA, NMR, 31 STRUCTURES | | Descriptor: | RNA 3UTR, U1A 102 | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1997-08-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the RNA-binding specificity of human U1A protein.
EMBO J., 16, 1997
|
|
2VIK
 
 | |
