8BZT
 
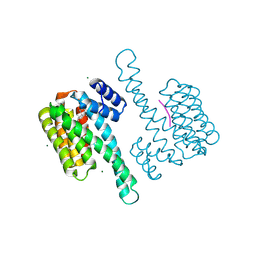 | | FC-NAg stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
7L1Y
 
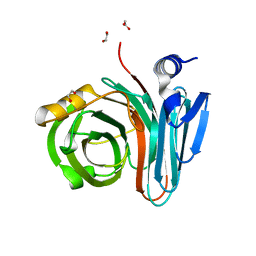 | | Unlocking the structural features for the exo-xylobiosidase activity of an unusual GH11 member identified in a compost-derived consortium-xylobiose complex | | Descriptor: | 1,2-ETHANEDIOL, Exo-B-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I, Briganti, L, Evangelista, D.E. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unlocking the structural features for the xylobiohydrolase activity of an unusual GH11 member identified in a compost-derived consortium.
Biotechnol.Bioeng., 118, 2021
|
|
8BZH
 
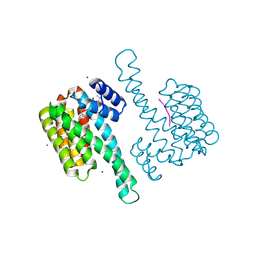 | | FC-NAc stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
8BZG
 
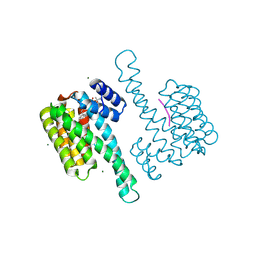 | | FC-31 stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin 31, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
5CGZ
 
 | | Crystal structure of GalB, the 4-carboxy-2-hydroxymuconate hydratase, from Pseuodomonas putida KT2440 | | Descriptor: | 4-oxalmesaconate hydratase, GLYCEROL, ZINC ION | | Authors: | Mazurkewich, S, Brott, A.S, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2015-07-09 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and Kinetic Characterization of the 4-Carboxy-2-hydroxymuconate Hydratase from the Gallate and Protocatechuate 4,5-Cleavage Pathways of Pseudomonas putida KT2440.
J.Biol.Chem., 291, 2016
|
|
6UQY
 
 | |
5XRC
 
 | |
8JNO
 
 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5MPS
 
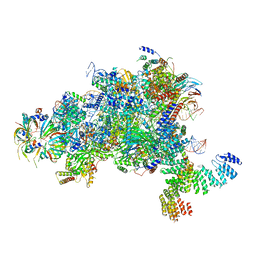 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5UGJ
 
 | |
8C0L
 
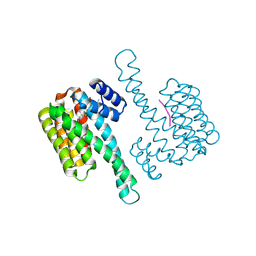 | | FC-THF stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin A-THF | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-17 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
7ZLT
 
 | |
5CHO
 
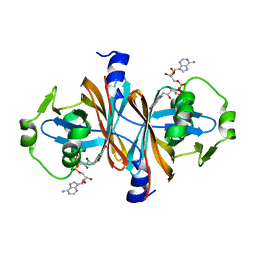 | |
8J3M
 
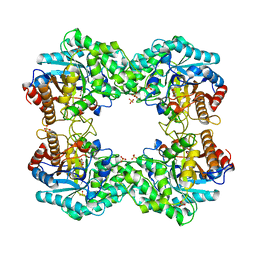 | |
8J5L
 
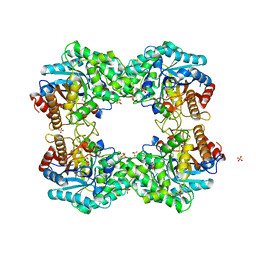 | |
5UHU
 
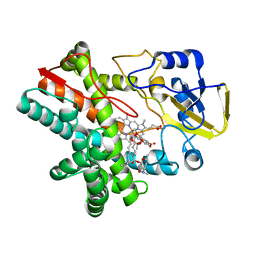 | |
6V37
 
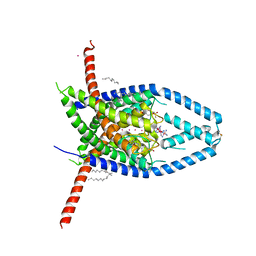 | |
7ZWA
 
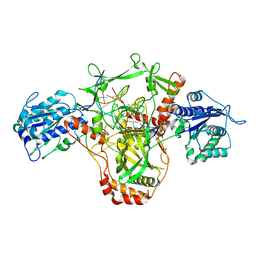 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8J5M
 
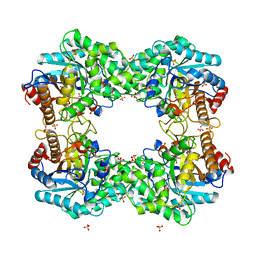 | |
6V3I
 
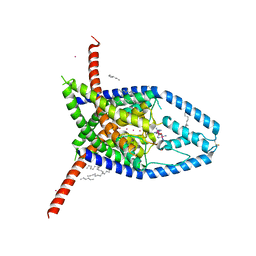 | |
5U97
 
 | | Crystal structure of Co-CAO1 in complex with piceatannol | | Descriptor: | BENZOIC ACID, COBALT (II) ION, Carotenoid oxygenase 1, ... | | Authors: | Sui, X, Palczewski, K, Banerjee, S, Kiser, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center.
Biochemistry, 56, 2017
|
|
8JB1
 
 | |
5CGC
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
1B2R
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
5CNG
 
 | | ultrafast dynamics in myoglobin: 150 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
