5VSI
 
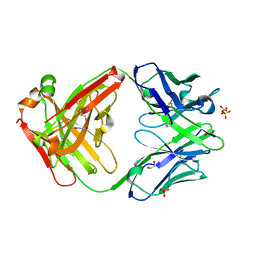 | | CH1/Ckappa Fab mutant 15.1 | | Descriptor: | 1,2-ETHANEDIOL, CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain, ... | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
4DYZ
 
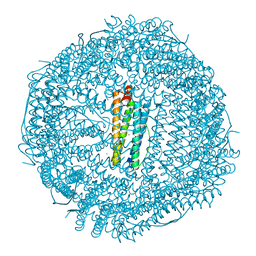 | |
4DZ0
 
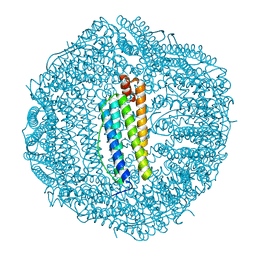 | |
2OFJ
 
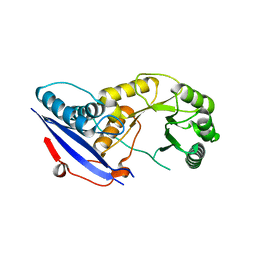 | | Crystal structure of the E190A mutant of o-succinylbenzoate synthase from Escherichia coli | | Descriptor: | O-succinylbenzoate synthase | | Authors: | Nagatani, R.A, Gonzalez, A, Shoichet, B.K, Brinen, L.S, Babbitt, P.C. | | Deposit date: | 2007-01-03 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stability for Function Trade-Offs in the Enolase Superfamily "Catalytic Module".
Biochemistry, 46, 2007
|
|
7T99
 
 | |
2A24
 
 | | HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain protein 1 | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-06-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization.
J.Mol.Biol., 353, 2005
|
|
2A7K
 
 | | carboxymethylproline synthase (CarB) from pectobacterium carotovora, apo enzyme | | Descriptor: | CarB | | Authors: | Sleeman, M.C, Sorensen, J.L, Batchelar, E.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Mechanistic Studies on Carboxymethylproline Synthase (CarB), a Unique Member of the Crotonase Superfamily Catalyzing the First Step in Carbapenem Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
7TZB
 
 | | Crystal structure of the human mitochondrial seryl-tRNA synthetase (mt SerRS) bound with a seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial | | Authors: | Kuhle, B, Hirschi, M, Doerfel, L, Lander, G, Schimmel, P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7U2B
 
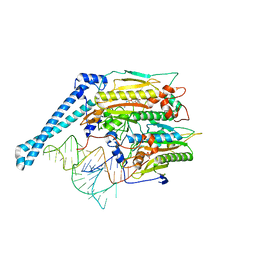 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
2A81
 
 | | carboxymethylproline synthase (CarB) from pectobacterium carotovora, complexed with acetyl CoA and bicine | | Descriptor: | ACETYL COENZYME *A, BICINE, CarB | | Authors: | Sleeman, M.C, Sorensen, J.L, Batchelar, E.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2005-07-07 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Mechanistic Studies on Carboxymethylproline Synthase (CarB), a Unique Member of the Crotonase Superfamily Catalyzing the First Step in Carbapenem Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
7U2A
 
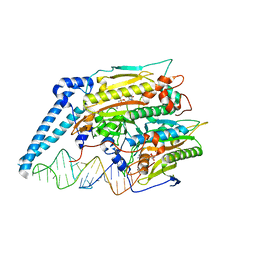 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (38-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
2JKS
 
 | | Crystal structure of the the bradyzoite specific antigen BSR4 from toxoplasma gondii. | | Descriptor: | BRADYZOITE SURFACE ANTIGEN BSR4, ZINC ION | | Authors: | Boulanger, M.J, Grigg, M.E, Bruic, E, Grujic, O. | | Deposit date: | 2008-08-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of the Bradyzoite Surface Antigen (Bsr4) from Toxoplasma Gondii, a Unique Addition to the Surface Antigen Glycoprotein 1-Related Superfamily.
J.Biol.Chem., 284, 2009
|
|
7T98
 
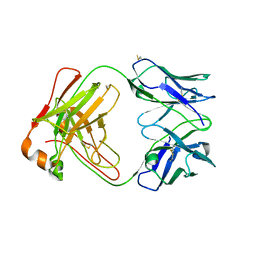 | |
7T97
 
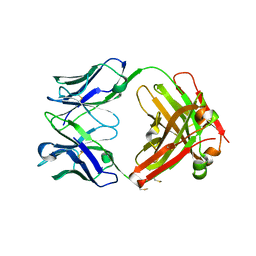 | |
3J6P
 
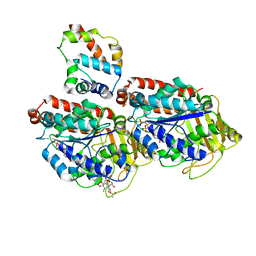 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | Descriptor: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
7VU5
 
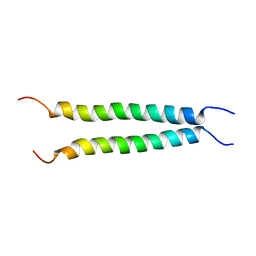 | |
1Z7N
 
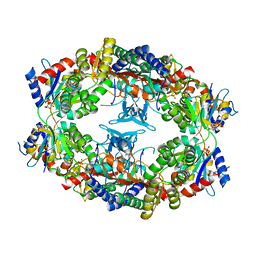 | | ATP Phosphoribosyl transferase (HisZG ATP-PRTase) from Lactococcus lactis with bound PRPP substrate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Champagne, K.S, Sissler, M, Larrabee, Y, Doublie, S, Francklyn, C.S. | | Deposit date: | 2005-03-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog.
J.Biol.Chem., 280, 2005
|
|
5A71
 
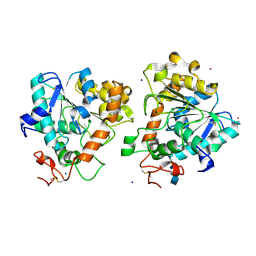 | | Open and closed conformations and protonation states of Candida antarctica Lipase B: atomic resolution native | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, LIPASE B, ... | | Authors: | Stauch, B, Fisher, S.J, Cianci, M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Open and Closed States of Candida Antarctica Lipase B: Protonation and the Mechanism of Interfacial Activation.
J.Lipid Res., 56, 2015
|
|
3HDD
 
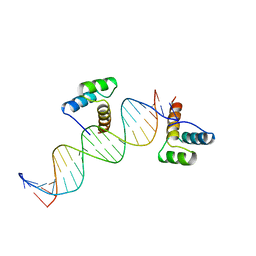 | | ENGRAILED HOMEODOMAIN DNA COMPLEX | | Descriptor: | 5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3', ENGRAILED HOMEODOMAIN | | Authors: | Fraenkel, E, Rould, M.A, Chambers, K.A, Pabo, C.O. | | Deposit date: | 1998-07-13 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures.
J.Mol.Biol., 284, 1998
|
|
1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
1Z7M
 
 | | ATP Phosphoribosyl transferase (HisZG ATP-PRTase) from Lactococcus lactis | | Descriptor: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, PHOSPHATE ION, ... | | Authors: | Champagne, K.S, Sissler, M, Larrabee, Y, Doublie, S, Francklyn, C.S. | | Deposit date: | 2005-03-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog.
J.Biol.Chem., 280, 2005
|
|
2KQM
 
 | |
2KQN
 
 | |
2KXA
 
 | |
