2W00
 
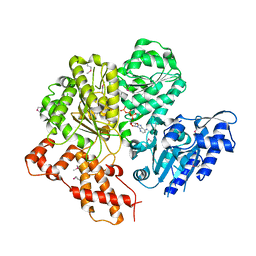 | | Crystal structure of the HsdR subunit of the EcoR124I restriction enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HSDR, MAGNESIUM ION | | Authors: | Lapkouski, M, Panjikar, S, Kuta Smatanova, I, Ettrich, R, Csefalvay, E. | | Deposit date: | 2008-08-08 | | Release date: | 2008-12-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Motor Subunit of Type I Restriction-Modification Complex Ecor124I.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1ASW
 
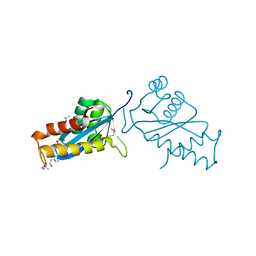 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 20% PEG 4000, 10% ISOPROPANOL, HEPES PH 7.5 USING SELENOMETHIONINE SUBSTITUTED PROTEIN; DATA COLLECTED AT-165 DEGREES C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, ISOPROPYL ALCOHOL | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
7UIC
 
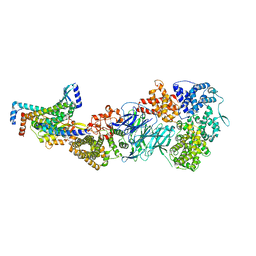 | | Mediator-PIC Early (Tail A) | | Descriptor: | Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 15, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7UIL
 
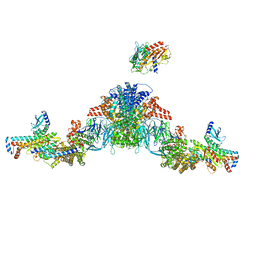 | | Mediator-PIC Early (Tail A/B Dimer) | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 15, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7YDT
 
 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
4OQ2
 
 | | 5hmC specific restriction endonuclease PvuRTs1I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Restriction endonuclease PvuRts1 I | | Authors: | Kazrani, A.A, Kowalska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the 5hmC specific endonuclease PvuRts1I.
Nucleic Acids Res., 42, 2014
|
|
3GCB
 
 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
1RI7
 
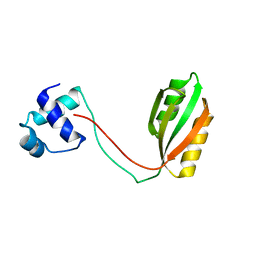 | |
1HQ3
 
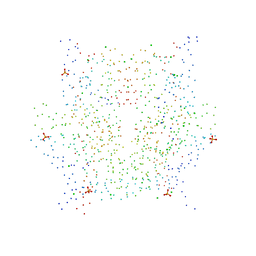 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HQC
 
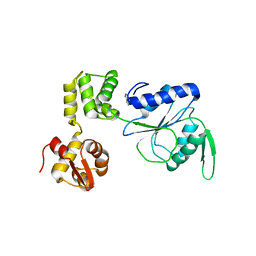 | | STRUCTURE OF RUVB FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | ADENINE, MAGNESIUM ION, RUVB | | Authors: | Yamada, K, Kunishima, N, Mayanagi, K, Iwasaki, H, Morikawa, K. | | Deposit date: | 2000-12-15 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1SKP
 
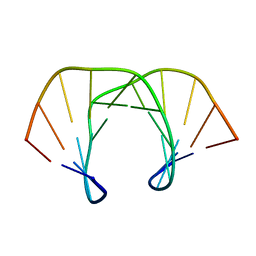 | | NMR STRUCTURE OF D(GCATATGATAG)(DOT)D(CTATCATATGC): A CONSENSUS SEQUENCE FOR PROMOTERS RECOGNIZED BY SIGMA-K RNA POLYMERASE, 4 STRUCTURES | | Descriptor: | SIGMA-K RNA POLYMERASE CONSENSUS SEQUENCE | | Authors: | Tonelli, M, Ragg, E, Bianucci, A.M, Lesiak, K, James, T.L. | | Deposit date: | 1998-05-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of d(GCATATGATAG). d(CTATCATATGC): a consensus sequence for promoters recognized by sigma K RNA polymerase.
Biochemistry, 37, 1998
|
|
1J0R
 
 | | Crystal structure of the replication termination protein mutant C110S | | Descriptor: | replication termination protein | | Authors: | Vivian, J.P, Hastings, A.F, Duggin, I.G, Wake, R.G, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2002-11-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The impact of single cysteine residue mutations on the replication terminator protein
Biochem.Biophys.Res.Commun., 310, 2003
|
|
6M2M
 
 | |
6S1R
 
 | |
1UFI
 
 | | Crystal structure of the dimerization domain of human CENP-B | | Descriptor: | Major centromere autoantigen B | | Authors: | Tawaramoto, M.S, Kurumizaka, H, Tanaka, Y, Park, S.-Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-30 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human centromere protein B (CENP-B) dimerization domain at 1.65-A resolution
J.Biol.Chem., 278, 2003
|
|
5XV9
 
 | |
3QMG
 
 | | Structural Basis of Selective Binding of Non-Methylated CpG islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*AP*CP*GP*GP*TP*GP*GP*C)-3', 5'-D(*GP*CP*CP*AP*CP*CP*GP*TP*TP*GP*GP*C)-3', CpG-binding protein, ... | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
4L9F
 
 | | Structure of a SeMet derivative of PpsR Q-PAS1 from Rb. sphaeroides | | Descriptor: | Transcriptional regulator, PpsR | | Authors: | Heintz, U, Meinhart, A, Schlichting, I, Winkler, A. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-12 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B0J
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1J9N
 
 | | Solution Structure of the Nucleopeptide [AC-LYS-TRP-LYS-HSE(p3*dGCATCG)-ALA]-[p5*dCGTAGC] | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*CP*(PGN))-3', peptide ACE-LYS-TRP-LYS-HSE-ALA | | Authors: | Gomez-Pinto, I, Marchan, V, Gago, F, Grandas, A, Gonzalez, C. | | Deposit date: | 2001-05-28 | | Release date: | 2003-01-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of tryptophan-containing nucleopeptide duplexes
Chembiochem, 4, 2003
|
|
5B0M
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-dodecyl maltoside | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7PDQ
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with LG100268 and a coactivator fragment | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
7PDT
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with BMS649 and a coactivator fragment | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
4LRZ
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase operon regulatory protein, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
