3E5U
 
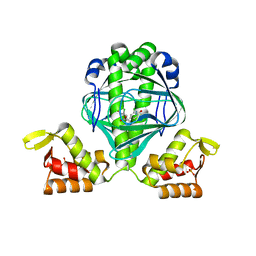 | | OCPA complexed CprK (C200S) | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein, PHOSPHATE ION, ... | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
2JTM
 
 | | Solution structure of Sso6901 from Sulfolobus solfataricus P2 | | Descriptor: | Putative uncharacterized protein | | Authors: | Feng, Y, Guo, L, Huang, L, Wang, J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea
Nucleic Acids Res., 36, 2008
|
|
1OVF
 
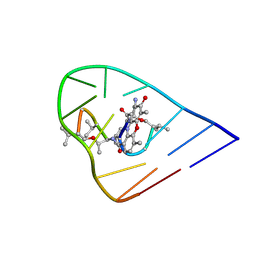 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
1EA6
 
 | |
2Z7C
 
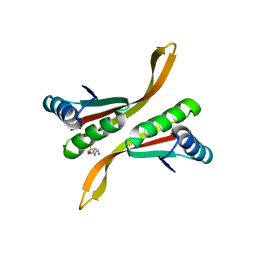 | | Crystal structure of chromatin protein alba from hyperthermophilic archaeon pyrococcus horikoshii | | Descriptor: | ARGININE, DNA/RNA-binding protein Alba | | Authors: | Hada, K, Nakashima, T, Osawa, T, Shimada, H, Kakuta, Y, Kimura, M. | | Deposit date: | 2007-08-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3.
Biosci.Biotechnol.Biochem., 72, 2008
|
|
4L5Q
 
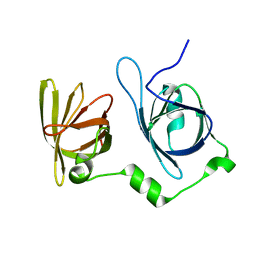 | | Crystal structure of p202 HIN1 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
2BNM
 
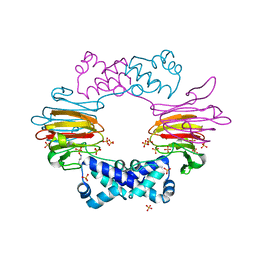 | |
5YJE
 
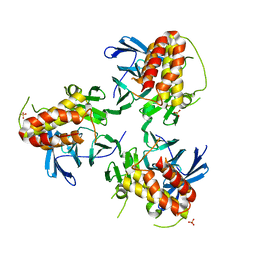 | | Crystal structure of HIRA(644-1017) | | Descriptor: | Protein HIRA, SULFATE ION | | Authors: | Sato, Y, Senda, M, Senda, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit
Nat Commun, 9, 2018
|
|
7T8L
 
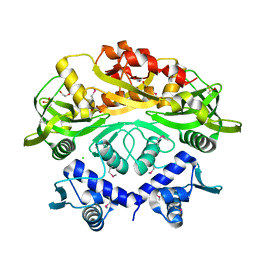 | | BrxR from Acinetobacter BREX type I phage restriction system | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
3DKX
 
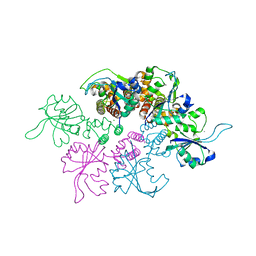 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3E0F
 
 | |
3DKY
 
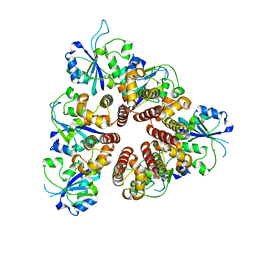 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
6SQW
 
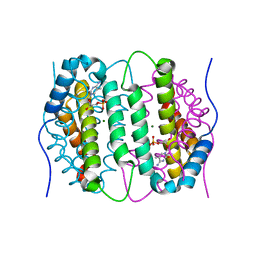 | | Mouse dCTPase in complex with 5-Me-dCMP | | Descriptor: | 5-METHYL-2'-DEOXY-CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
2BNO
 
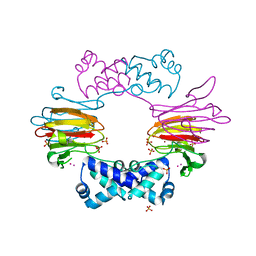 | | The structure of Hydroxypropylphosphonic acid epoxidase from S. wedmorenis. | | Descriptor: | EPOXIDASE, MERCURY (II) ION, SULFATE ION, ... | | Authors: | McLuskey, K, Cameron, S, Hunter, W.N. | | Deposit date: | 2005-03-29 | | Release date: | 2005-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Reactivity of Hydroxypropylphosphonic Acid Epoxidase in Fosfomycin Biosynthesis by a Cation- and Flavin-Dependent Mechanism.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1L3M
 
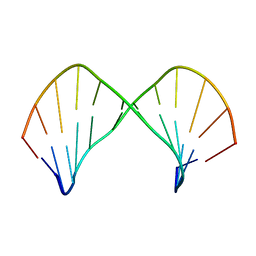 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
8V44
 
 | |
2ZIU
 
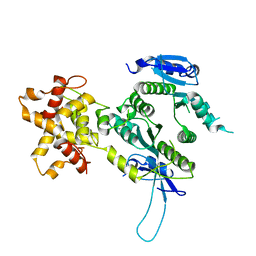 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
1LJ9
 
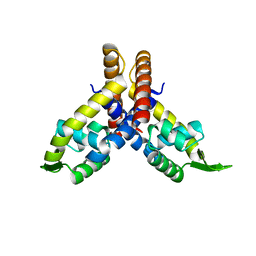 | | The crystal structure of the transcriptional regulator SlyA | | Descriptor: | transcriptional regulator SlyA | | Authors: | Wu, R.Y, Zhang, R.G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-04-19 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor
J.Biol.Chem., 278, 2003
|
|
2BNN
 
 | |
6SQZ
 
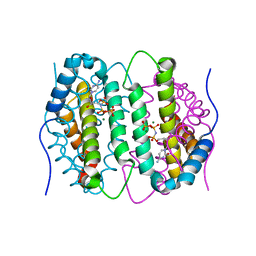 | | Mouse dCTPase in complex with dCMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
2ZIV
 
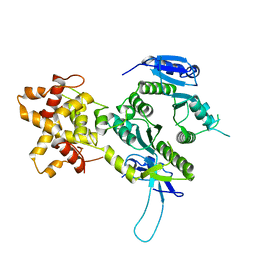 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
1GHT
 
 | |
2ZIX
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
8HX0
 
 | |
4KHA
 
 | | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Histone H2A, ... | | Authors: | Hondele, M, Halbach, F, Hassler, M, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
