5R91
 
 | |
2M17
 
 | |
4G29
 
 | |
4AL2
 
 | |
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
4CHE
 
 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4DX7
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DECANE, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3G9W
 
 | | Crystal Structure of Talin2 F2-F3 in Complex with the Integrin Beta1D Cytoplasmic Tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Integrin beta-1D, ... | | Authors: | Anthis, N.J, Wegener, K.L, Ye, F, Kim, C, Lowe, E.D, Vakonakis, I, Bate, N, Critchley, D.R, Ginsberg, M.H, Campbell, I.D. | | Deposit date: | 2009-02-15 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | The structure of an integrin/talin complex reveals the basis of inside-out signal transduction
Embo J., 28, 2009
|
|
7A24
 
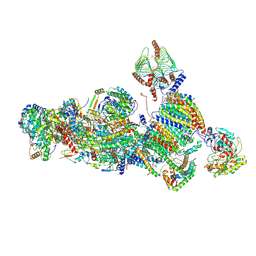 | | Assembly intermediate of the plant mitochondrial complex I | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 13kDa, 15kDa, ... | | Authors: | Soufari, H, Waltz, F, Hashem, Y. | | Deposit date: | 2020-08-16 | | Release date: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM.
Nat Commun, 11, 2020
|
|
2HPI
 
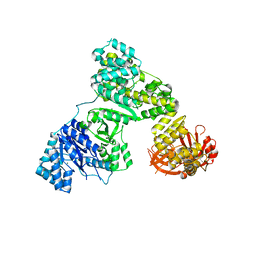 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HRK
 
 | |
2NPB
 
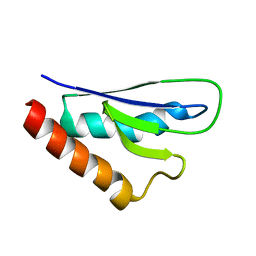 | | NMR solution structure of mouse SelW | | Descriptor: | Selenoprotein W | | Authors: | Aachmann, F.L, Fomenko, D.E, Soragni, A, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins
J.Biol.Chem., 282, 2007
|
|
3AOA
 
 | |
2J4W
 
 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
3AOB
 
 | |
2YKH
 
 | | Sensor region of a sensor histidine kinase | | Descriptor: | PROBABLE SENSOR HISTIDINE KINASE PDTAS | | Authors: | Preu, J, Panjikar, S, Morth, P, Jaiswal, R, Karunakar, P, Tucker, P.A. | | Deposit date: | 2011-05-27 | | Release date: | 2012-06-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Sensor Region of the Ubiquitous Cytosolic Sensor Kinase, Pdtas, Contains Pas and Gaf Domain Sensing Modules.
J.Struct.Biol., 177, 2012
|
|
2FN1
 
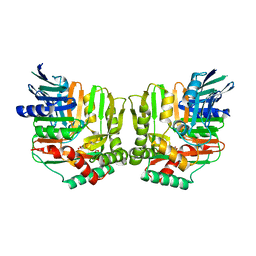 | | Crystal structures of Yersinia enterocolitica salicylate synthase (Irp9) in complex with the reaction products salicylate and pyruvate | | Descriptor: | 2-HYDROXYBENZOIC ACID, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kerbarh, O, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2006-01-10 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Yersinia enterocolitica Salicylate Synthase and its Complex with the Reaction Products Salicylate and Pyruvate.
J.Mol.Biol., 357, 2006
|
|
6Y11
 
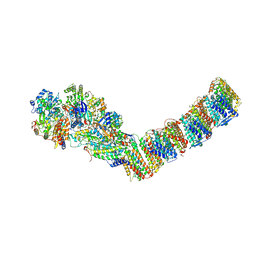 | | Respiratory complex I from Thermus thermophilus | | Descriptor: | DUF3197 domain-containing protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
4FBL
 
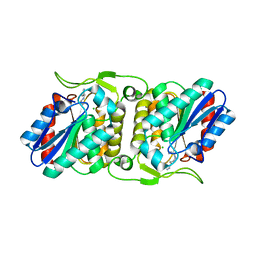 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
2J5L
 
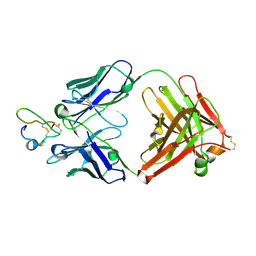 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
4G2B
 
 | |
4K7Q
 
 | | Crystal Structure of AcrB Complexed with Linezolid at 3.5 Resolution | | Descriptor: | Acriflavine resistance protein B, N-{[(5S)-3-(3-fluoro-4-morpholin-4-ylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide | | Authors: | Hung, L.W, Kim, H.B, Murakami, S, Gupta, G, Kim, C.Y, Terwilliger, T.C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB complexed with linezolid at 3.5 Angstrom resolution.
J.Struct.Funct.Genom., 14, 2013
|
|
6C94
 
 | |
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
3HZU
 
 | |
