3GQ0
 
 | |
3GQA
 
 | |
3GW0
 
 | | UROD mutant G318R | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3R5H
 
 | | Crystal Structure of ADP-AIR complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3GR2
 
 | |
3GWU
 
 | | Leucine transporter LeuT in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, LEUCINE, SODIUM ION, ... | | Authors: | Zhou, Z, Zhen, J, Karpowich, N.K, Law, C.J, Reith, M.E.A, Wang, D.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GRD
 
 | |
3QGF
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QLZ
 
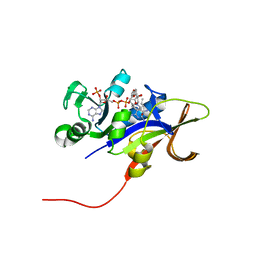 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine (UCP130B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
3GXF
 
 | |
3QMB
 
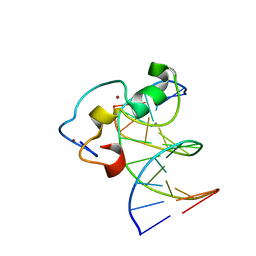 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3', CALCIUM ION, CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3GSW
 
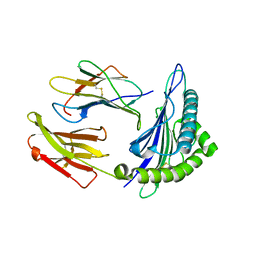 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-T8A peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant T8A (NLVPMVAAV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3QIA
 
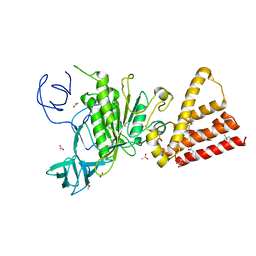 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3GZ4
 
 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3QNQ
 
 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
3QJ8
 
 | |
3GZI
 
 | |
3GUF
 
 | |
3GXB
 
 | | Crystal structure of VWF A2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Springer, T.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GXO
 
 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | Descriptor: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3QR0
 
 | | Crystal Structure of S. officinalis PLC21 | | Descriptor: | CALCIUM ION, GLYCEROL, phospholipase C-beta (PLC-beta) | | Authors: | Lyon, A.M, Northup, J.K, Tesmer, J.J.G. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An autoinhibitory helix in the C-terminal region of phospholipase C-beta mediates Galphaq activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QRR
 
 | | Structure of Thermus Thermophilus Cse3 bound to an RNA representing a product complex | | Descriptor: | Putative uncharacterized protein TTHB192, RNA (5'-R(*GP*UP*CP*CP*CP*CP*AP*CP*GP*CP*GP*UP*GP*UP*GP*GP*GP*(23G))-3') | | Authors: | Schellenberg, M.J, Gesner, E.G, Garside, E.L, MacMillan, A.M. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Recognition and maturation of effector RNAs in a CRISPR interference pathway.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3H0N
 
 | |
3QMQ
 
 | | Crystal Structure of E. coli LsrG | | Descriptor: | Autoinducer-2 (AI-2) modifying protein LsrG | | Authors: | Miller, S.T, Russell, C. | | Deposit date: | 2011-02-04 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processing the Interspecies Quorum-sensing Signal Autoinducer-2 (AI-2): CHARACTERIZATION OF PHOSPHO-(S)-4,5-DIHYDROXY-2,3-PENTANEDIONE ISOMERIZATION BY LsrG PROTEIN.
J.Biol.Chem., 286, 2011
|
|
3H0V
 
 | | Human AdoMetDC with 5'-Deoxy-5'-(dimethylsulfonio) adenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-deoxy-5'-(dimethyl-lambda~4~-sulfanyl)adenosine, PYRUVIC ACID, ... | | Authors: | Bale, S, Brooks, W.H, Hanes, J.W, Mahesan, A.M, Guida, W.C, Ealick, S.E. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Role of the sulfonium center in determining the ligand specificity of human s-adenosylmethionine decarboxylase.
Biochemistry, 48, 2009
|
|
