6UE8
 
 | | Structure of tetrameric sIgA complex (Class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
7V3T
 
 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
4BP9
 
 | | Oligopeptidase B from Trypanosoma brucei with covalently bound antipain - closed form | | Descriptor: | ANTIPAIN, OLIGOPEPTIDASSE B | | Authors: | Canning, P, Rea, D, Morty, R, Fulop, V. | | Deposit date: | 2013-05-23 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of Trypanosoma Brucei Oligopeptidase B Broaden the Paradigm of Catalytic Regulation in Prolyl Oligopeptidase Family Enzymes.
Plos One, 8, 2013
|
|
4X3X
 
 | | The crystal structure of Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4WNZ
 
 | |
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7NJF
 
 | | Hen egg white lysozyme (HEWL) grown inside HARE serial crystallography chip | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJJ
 
 | | Proteinase K grown inside HARE serial crystallography chip | | Descriptor: | NITRATE ION, Proteinase K | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NKF
 
 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJE
 
 | | gamma(S)-crystallin 9-site deamidation mutant grown inside HARE serial crystallography chip | | Descriptor: | Gamma-crystallin S | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJH
 
 | | HEX1 (in cellulo) grown inside HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJG
 
 | | Xylose isomerase grown inside HARE serial crystallography chip | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJI
 
 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7UNX
 
 | |
6SO0
 
 | | NMR solution structure of the family 14 carbohydrate binding module (CBM14) from human chitotriosidase | | Descriptor: | Chitotriosidase-1 | | Authors: | Madland, E, Crasson, O, Vandevenne, M, Sorlie, M, Aachmann, F.L. | | Deposit date: | 2019-08-28 | | Release date: | 2020-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase.
Acs Omega, 4, 2019
|
|
4AQB
 
 | | MBL-Ficolin Associated Protein-1, MAP-1 aka MAP44 | | Descriptor: | CALCIUM ION, MANNAN-BINDING LECTIN SERINE PROTEASE 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skjoedt, M.O, Roversi, P, Hummelshoj, T, Palarasah, Y, Johnson, S, Lea, S.M, Garred, P. | | Deposit date: | 2012-04-16 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure and Functional Characterization of the Complement Regulator Mannose-Binding Lectin (Mbl)/Ficolin-Associated Protein-1 (Map-1).
J.Biol.Chem., 287, 2012
|
|
7TRA
 
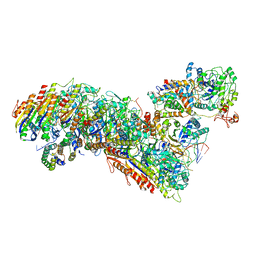 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
6FDT
 
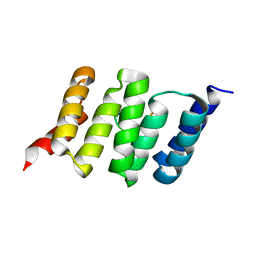 | |
8GC5
 
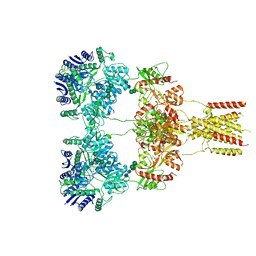 | | Domoate-bound GluK2 kainate receptors in non-active conformation | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC3
 
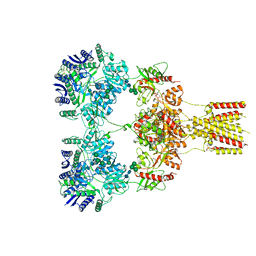 | | Domote-bound GluK2 kainate receptors in partially-open conformation 2 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC4
 
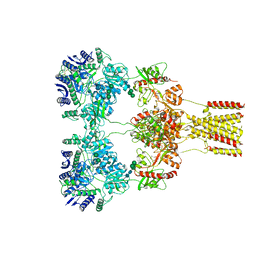 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 3 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
7MZX
 
 | |
4B8S
 
 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-DEPENDENT GLUCOKINASE, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
6T72
 
 | | Structure of the RsaA N-terminal domain bound to LPS | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | von Kuegelgen, A, Bharat, T.A.M. | | Deposit date: | 2019-10-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
3ZCS
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(1-naphthoyl) -N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
