1C30
 
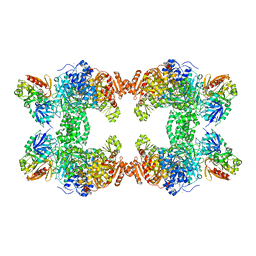 | | CRYSTAL STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTATION C269S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-24 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
4N99
 
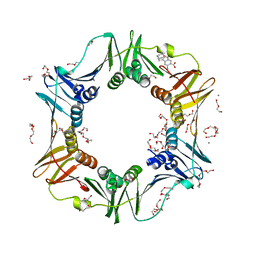 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
1A9X
 
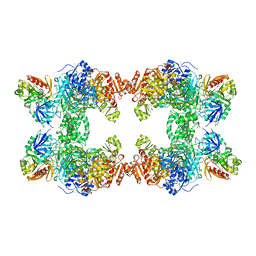 | |
1DED
 
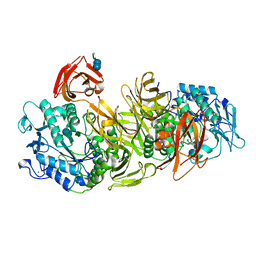 | | CRYSTAL STRUCTURE OF ALKALOPHILIC ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE COMPLEXED WITH AN INHIBITOR, ACARBOSE, AT 2.0 A RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-11-14 | | Release date: | 2000-04-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alkalophilic asparagine 233-replaced cyclodextrin glucanotransferase complexed with an inhibitor, acarbose, at 2.0 A resolution.
J.Biochem.(Tokyo), 127, 2000
|
|
1CS0
 
 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
2ECP
 
 | | THE CRYSTAL STRUCTURE OF THE E. COLI MALTODEXTRIN PHOSPHORYLASE COMPLEX | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | O'Reilly, M, Watson, K.A, Johnson, L.N. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of the Escherichia coli maltodextrin phosphorylase-acarbose complex.
Biochemistry, 38, 1999
|
|
3PFD
 
 | |
2F6D
 
 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
8YIE
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
6WL9
 
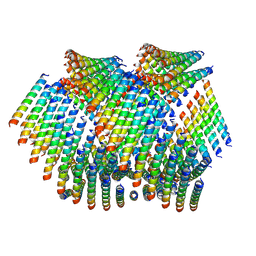 | | Cryo-EM of Form 2 like peptide filament, Form2a | | Descriptor: | peptide Form2a | | Authors: | Wang, F, Beltran, L.C, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WXE
 
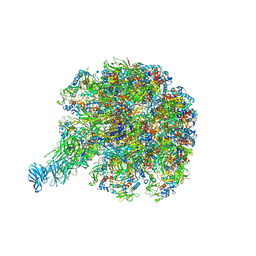 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WFT
 
 | |
6WVV
 
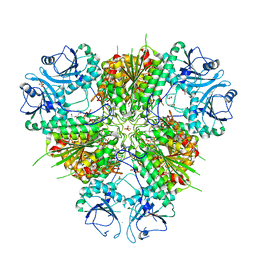 | | Plasmodium vivax M17 leucyl aminopeptidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, M17 leucyl aminopeptidase, ... | | Authors: | Malcolm, T.R, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
6WH3
 
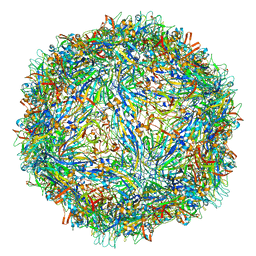 | | Capsid structure of Penaeus monodon metallodensovirus at pH 8.2 | | Descriptor: | CALCIUM ION, Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | Descriptor: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | Authors: | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
4ZX8
 
 | |
4ZYQ
 
 | |
6WEO
 
 | | IL-22 Signaling Complex with IL-22R1 and IL-10Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saxton, R.A, Jude, K.M, Henneberg, L.T, Garcia, K.C. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design.
Immunity, 54, 2021
|
|
6WKV
 
 | | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE) | | Descriptor: | Encapsulin, FLAVIN MONONUCLEOTIDE | | Authors: | Williams, E, Jenkins, M, Zhao, H, Juneja, P, Lutz, S. | | Deposit date: | 2020-04-17 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE)
To Be Published
|
|
4ZY0
 
 | |
4ZY2
 
 | | X-ray crystal structure of PfA-M17 in complex with hydroxamic acid-based inhibitor 10o | | Descriptor: | CARBONATE ION, DIMETHYL SULFOXIDE, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluorobiphenyl-4-yl)ethyl]-2,2-dimethylpropanamide, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-21 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent dual inhibitors of Plasmodium falciparum M1 and M17 aminopeptidases through optimization of S1 pocket interactions.
Eur.J.Med.Chem., 110, 2016
|
|
4ZX9
 
 | |
5AJ3
 
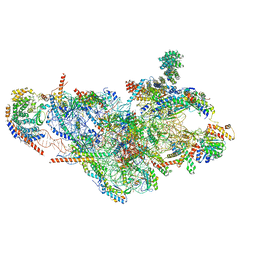 | | Structure of the small subunit of the mammalian mitoribosome | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MITORIBOSOMAL 12S RRNA, ... | | Authors: | Greber, B.J, Bieri, P, Leibundgut, M, Leitner, A, Aebersold, R, Boehringer, D, Ban, N. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome.
Science, 348, 2015
|
|
7AZ5
 
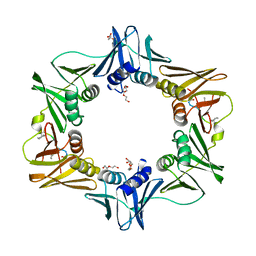 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
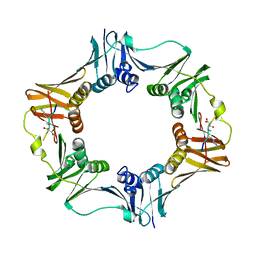 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
