7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
8DFL
 
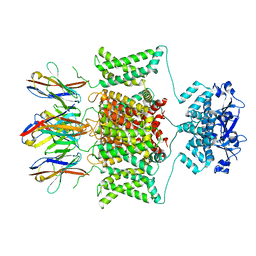 | |
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSZ
 
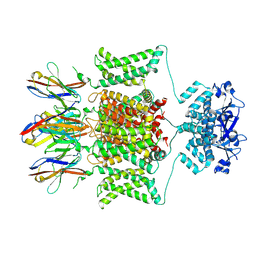 | | Structure of human Kv1.3 with A0194009G09 nanobodies | | Descriptor: | Nanobody A0194009G09, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSY
 
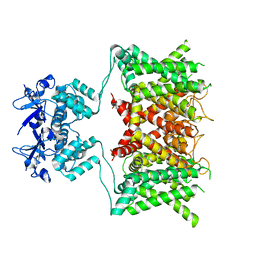 | | Structure of human Kv1.3 (alternate conformation) | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSV
 
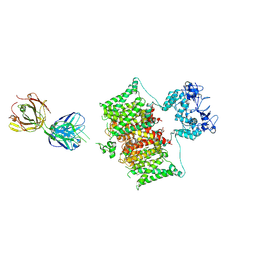 | | Structure of human Kv1.3 with Fab-ShK fusion | | Descriptor: | Fab-ShK fusion, heavy chain, light chain, ... | | Authors: | Meyerson, J.R, Selvakumar, P, Smider, V, Huang, R. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7EYE
 
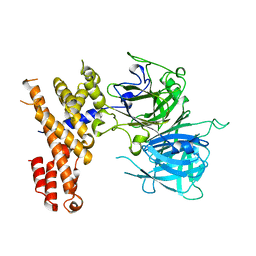 | |
7EYQ
 
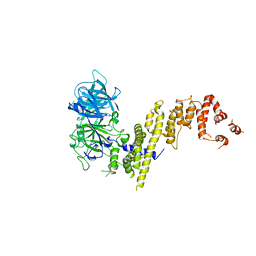 | |
7X5V
 
 | |
7SQY
 
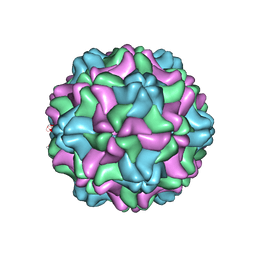 | | CSDaV GFP mutant | | Descriptor: | Citrus Sudden Death-associated Virus Capsid Protein,Green fluorescent protein,Citrus Sudden Death-associated Virus Capsid Protein | | Authors: | Guo, F, Matsumura, E.E, Falk, B.W. | | Deposit date: | 2021-11-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Citrus sudden death-associated virus as a new expression vector for rapid in planta production of heterologous proteins, chimeric virions, and virus-like particles.
Biotechnol Rep., 35, 2022
|
|
7Z7P
 
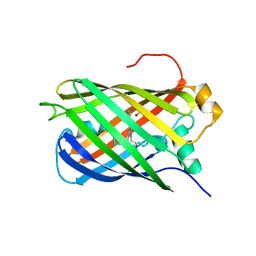 | |
7Z7Q
 
 | |
7Z7O
 
 | |
7SWS
 
 | | Crystal structure of the chromoprotein amilCP | | Descriptor: | BROMIDE ION, CHLORIDE ION, Chromoprotein amilCP | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWT
 
 | | Crystal structure of the chromoprotein eforRED | | Descriptor: | Chromoprotein eforRED | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWR
 
 | | Crystal structure of the chromoprotein gfasPurple | | Descriptor: | CHLORIDE ION, Chromoprotein gfasPurple | | Authors: | Caputo, A.T, Newman, J, Peat, T.S, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWU
 
 | | Crystal structure of the chromoprotein spisPINK | | Descriptor: | Chromoprotein spisPINK | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AMU
 
 | | Crystal structure of rsEGFP2 T204A in its fluorescent on-state | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Moreno-Chicano, T, Schlichting, I, Hartmann, E, Zala, N, Colletier, J.-P, Weik, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of rsEGFP2 in its fluorescent on-state at pH 8.0
To Be Published
|
|
7SF9
 
 | |
7SFA
 
 | |
7QGK
 
 | | The mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer in its red state | | Descriptor: | MAGNESIUM ION, The red form of the mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Gaivoronskii, F.A, Vlaskina, A.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer.
Int J Mol Sci, 23, 2022
|
|
7BHG
 
 | |
7PCZ
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, GLYCEROL, Green fluorescent protein | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PCA
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, FORMAMIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PD0
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Green fluorescent protein, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
