6PM9
 
 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
8KBH
 
 | | Structure of CbTad1 complexed with 2',3'-cGAMP and cA3 | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, Thoeris anti-defense 1, cGAMP | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
6YE3
 
 | | IL-2 in complex with a Fab fragment from UFKA-20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chains: A,D,G, Chains: B,E,H, ... | | Authors: | Karakus, U, Mittl, P, Boyman, O. | | Deposit date: | 2020-03-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Receptor-gated IL-2 delivery by an anti-human IL-2 antibody activates regulatory T cells in three different species.
Sci Transl Med, 12, 2020
|
|
6VFT
 
 | | Crystal structure of human delta protocadherin 17 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
6PN4
 
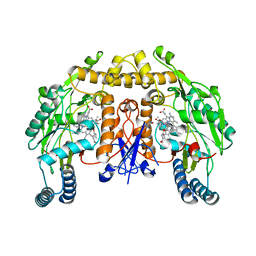 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclopropylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5V31
 
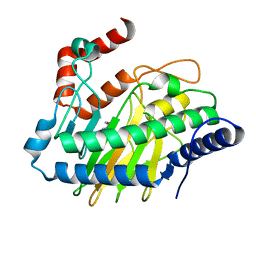 | | Ethylene forming enzyme in complex with manganese and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
6PNF
 
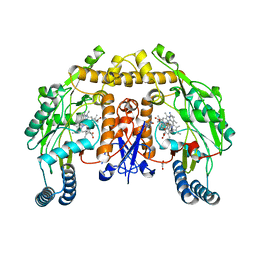 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6ML5
 
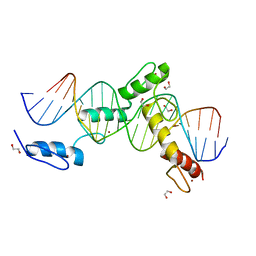 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6VGO
 
 | |
6POC
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(oxazol-4-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 7-{3-(aminomethyl)-4-[(1,3-oxazol-4-yl)methoxy]phenyl}-4-methylquinolin-2-amine, GLYCEROL, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5DHI
 
 | |
6G9E
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis - TsES1 - 6 molecules in ASU | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
7C8X
 
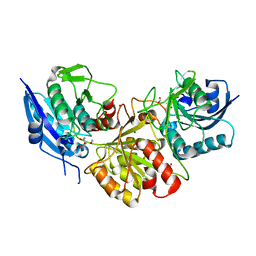 | | Blasnase-T13A with L-asn | | Descriptor: | ASPARAGINE, Asparaginase, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
5KVC
 
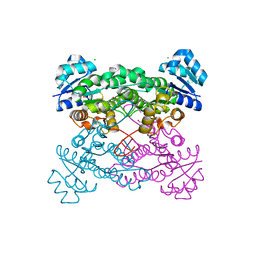 | |
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6POW
 
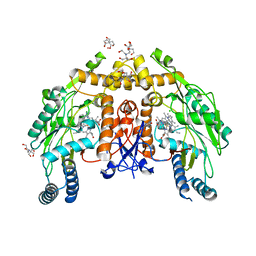 | | Structure of human endotheial nitric oxide synthase heme domain in complex with 7-(5-(Aminomethyl)pyridin-3-yl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[5-(aminomethyl)pyridin-3-yl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6Y1N
 
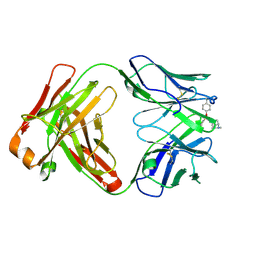 | | Crystal structure of the phosphonate-modified A.5 antibody FAB fragment | | Descriptor: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, FAB A.5 Heavy chain, FAB A.5 Light Chain | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GAQ
 
 | | Crystal structure of oxidised Flavodoxin 2 from Bacillus cereus | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin, ... | | Authors: | Lofstad, M, Gudim, I, Hersleth, H.-P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Characterization of Different Flavodoxin Reductase-Flavodoxin (FNR-Fld) Interactions Reveals an Efficient FNR-Fld Redox Pair and Identifies a Novel FNR Subclass.
Biochemistry, 57, 2018
|
|
5VQ9
 
 | | Structure of human TRIP13, Apo form | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5KWW
 
 | | Crystal Structure of Inhibitor JNJ-53718678 In Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[5-chloranyl-1-(3-methylsulfonylpropyl)indol-2-yl]methyl]-1-[2,2,2-tris(fluoranyl)ethyl]imidazo[4,5-c]pyridin-2-one, ... | | Authors: | McLellan, J.S, Battles, M.B, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2016-07-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor.
Nat Commun, 8, 2017
|
|
5HFR
 
 | | Crystal structure of the second bromodomain H395R mutant of human BRD3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, NITRATE ION | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Fonseca, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the second bromodomain H395R mutant of human BRD3
To Be Published
|
|
6PPQ
 
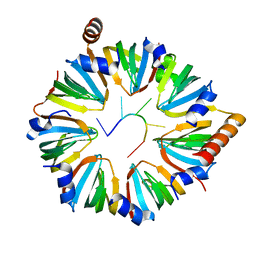 | |
6GB7
 
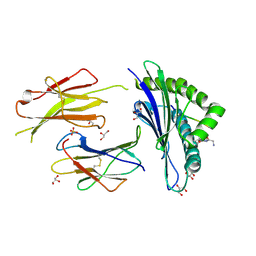 | | Structure of H-2Db with scoop loop from tapasin | | Descriptor: | Beta-2-microglobulin, GLY-GLY-LEU-SER, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5HX6
 
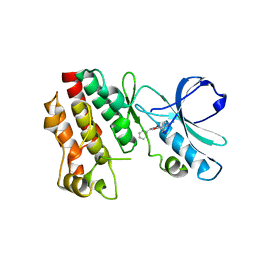 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6VDT
 
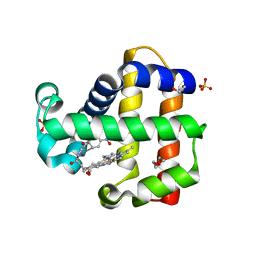 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Deuteroporphyrin IX and Substrate 4-nitrophenol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
